Purification and characterization of the lipid A 1-phosphatase LpxE of Rhizobium leguminosarum
- PMID: 18984595
- PMCID: PMC2610509
- DOI: 10.1074/jbc.M808390200
Purification and characterization of the lipid A 1-phosphatase LpxE of Rhizobium leguminosarum
Abstract
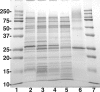
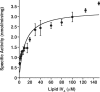
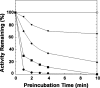
LpxE, a membrane-bound phosphatase found in Rhizobium leguminosarum and some other Gram-negative bacteria, selectively dephosphorylates the 1-position of lipid A on the outer surface of the inner membrane. LpxE belongs to the family of lipid phosphate phosphatases that contain a tripartite active site motif and six predicted transmembrane helices. Here we report the purification and characterization of R. leguminosarum LpxE. A modified lpxE gene, encoding a protein with an N-terminal His6 tag, was expressed in Escherichia coli. The protein was solubilized with Triton X-100 and purified to near-homogeneity. Gel electrophoresis reveals a molecular weight consistent with the predicted 31 kDa. LpxE activity is dependent upon Triton X-100, optimal near pH 6.5, and Mg2+-independent. The H197A and R133A substitutions inactivate LpxE, as does treatment with diethyl pyrocarbonate. In a mixed micelle assay system, the apparent Km for the precursor lipid IV(A) is 11 microm. Substrates containing the 3-deoxy-d-manno-oct-2-ulosonic acid disaccharide are dephosphorylated at similar rates to lipid IV(A), whereas glycerophospholipids like phosphatidic acid or phosphatidylglycerol phosphate are very poor substrates. However, an LpxE homologue present in Agrobacterium tumefaciens is selective for phosphatidylglycerol phosphate, demonstrating the importance of determining substrate specificity before assigning the functions of LpxE-related proteins. The availability of purified LpxE will facilitate the preparation of novel 1-dephosphorylated lipid A molecules that are not readily accessible by chemical methods.
Figures








References
-
- Fahy, E., Subramaniam, S., Brown, H. A., Glass, C. K., Merrill, A. H., Jr., Murphy, R. C., Raetz, C. R. H., Russell, D. W., Seyama, Y., Shaw, W., Shimizu, T., Spener, F., van Meer, G., VanNieuwenhze, M. S., White, S. H., Witztum, J. L., and Dennis, E. A. (2005) J. Lipid Res. 46 839-862 - PubMed
-
- Galloway, S. M., and Raetz, C. R. H. (1990) J. Biol. Chem. 265 6394-6402 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

