A one pot, one step, precision cloning method with high throughput capability
- PMID: 18985154
- PMCID: PMC2574415
- DOI: 10.1371/journal.pone.0003647
A one pot, one step, precision cloning method with high throughput capability
Abstract
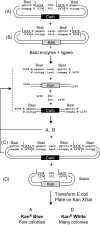
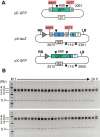
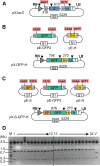
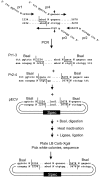
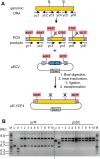
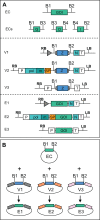
Current cloning technologies based on site-specific recombination are efficient, simple to use, and flexible, but have the drawback of leaving recombination site sequences in the final construct, adding an extra 8 to 13 amino acids to the expressed protein. We have devised a simple and rapid subcloning strategy to transfer any DNA fragment of interest from an entry clone into an expression vector, without this shortcoming. The strategy is based on the use of type IIs restriction enzymes, which cut outside of their recognition sequence. With proper design of the cleavage sites, two fragments cut by type IIs restriction enzymes can be ligated into a product lacking the original restriction site. Based on this property, a cloning strategy called 'Golden Gate' cloning was devised that allows to obtain in one tube and one step close to one hundred percent correct recombinant plasmids after just a 5 minute restriction-ligation. This method is therefore as efficient as currently used recombination-based cloning technologies but yields recombinant plasmids that do not contain unwanted sequences in the final construct, thus providing precision for this fundamental process of genetic manipulation.
Conflict of interest statement
Figures
References
-
- Marsischky G, LaBaer J. Many paths to many clones: a comparative look at high-throughput cloning methods. Genome Research. 2004;14:2020–2028. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

