A new theory of MHC evolution: beyond selection on the immune genes
- PMID: 18986972
- PMCID: PMC2660941
- DOI: 10.1098/rspb.2008.1299
A new theory of MHC evolution: beyond selection on the immune genes
Abstract
The major histocompatibility complex (MHC) is a dense region of immune genes with high levels of polymorphism, which are arranged in haplotype blocks. Traditional models of balancing selection (i.e. overdominance and negative frequency dependence) were developed to study the population genetics of single genes. However, the MHC is a multigene family surrounded by linked (non-neutral) polymorphisms, and not all of its features are well explained by these models. For example, (i) the high levels of polymorphism in small populations, (ii) the unexpectedly large genetic differentiation between populations, (iii) the shape of the allelic genealogy associated with trans-species evolution, and (iv) the close associations between particular MHC (human leucocyte antigen, HLA) haplotypes and the approximately 100 pathologies in humans. Here, I propose a new model of MHC evolution named Associative Balancing Complex evolution that can explain these phenomena. The model proposes that recessive deleterious mutations accumulate as a 'sheltered load' nearby MHC genes. These mutations can accumulate because (i) they are rarely expressed as homozygotes given the high MHC gene diversity and (ii) purifying selection is inefficient with low recombination rates (cf. Muller's ratchet). Once fixed, these mutations add to balancing selection and further reinforce linkage through epistatic selection against recombinants.
Figures


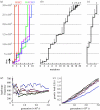
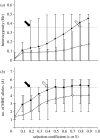
References
-
- Aguilar A., Roemer G., Debenham S., Binns M., Garcelon D., Wayne R.K. High MHC diversity maintained by balancing selection in an otherwise genetically monomorphic mammal. Proc. Natl Acad. Sci. USA. 2004;101:3490–3494. doi:10.1073/pnas.0306582101 - DOI - PMC - PubMed
-
- Bell G. University of California Press; Berkeley, CA: 1982. The masterpiece of nature: the evolution and genetics of sexuality.
-
- Bernatchez L., Landry C. MHC studies in nonmodel vertebrates: what have we learned about natural selection in 15 years? J. Evol. Biol. 2003;16:363–377. doi:10.1046/j.1420-9101.2003.00531.x - DOI - PubMed
-
- Bos D.H., Waldman B. Evolution by recombination and transspecies polymorphism in the MHC class I gene of Xenopus laevis. Mol. Biol. Evol. 2006;23:137–143. doi:10.1093/molbev/msj016 - DOI - PubMed
-
- Charlesworth D., Kamau E., Hagenblad J., Tang C.L. Trans-specificity at loci near the self-incompatibility loci in Arabidopsis. Genetics. 2006;172:2699–2704. doi:10.1534/genetics.105.051938 - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
