MicroRNA-298 and microRNA-328 regulate expression of mouse beta-amyloid precursor protein-converting enzyme 1
- PMID: 18986979
- PMCID: PMC2908704
- DOI: 10.1074/jbc.M807530200
MicroRNA-298 and microRNA-328 regulate expression of mouse beta-amyloid precursor protein-converting enzyme 1
Abstract
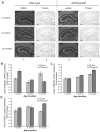
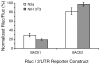
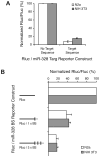
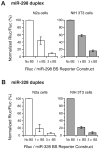
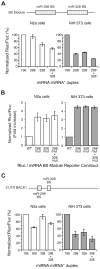
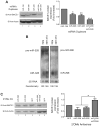
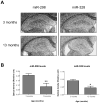
MicroRNAs (miRNAs) are key regulatory RNAs known to repress mRNA translation through recognition of specific binding sites located mainly in their 3'-untranslated region (UTR). Loss of specific miRNA control of gene expression is thus expected to underlie serious genetic diseases. Intriguingly, previous post-mortem analyses showed higher beta-amyloid precursor protein-converting enzyme (BACE) protein, but not mRNA, levels in the brain of patients that suffered from Alzheimer disease (AD). Here we also observed a loss of correlation between BACE1 mRNA and protein levels in the hippocampus of a mouse model of AD. Consistent with an impairment of miRNA-mediated regulation of BACE1 expression, these findings prompted us to investigate the regulatory role of the BACE1 3'-UTR element and the possible involvement of specific miRNAs in cultured neuronal (N2a) and fibroblastic (NIH 3T3) cells. Through various experimental approaches, we validated computational predictions and demonstrated that miR-298 and miR-328 recognize specific binding sites in the 3'-UTR of BACE1 mRNA and exert regulatory effects on BACE1 protein expression in cultured neuronal cells. Our results may provide the molecular basis underlying BACE1 deregulation in AD and offer new perspectives on the etiology of this neurological disorder.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

