Native R-loops persist throughout the mouse mitochondrial DNA genome
- PMID: 18986989
- PMCID: PMC2605977
- DOI: 10.1074/jbc.M806174200
Native R-loops persist throughout the mouse mitochondrial DNA genome
Abstract
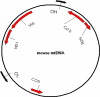
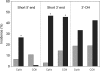
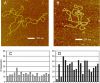
Mammalian mtDNA has been found here to harbor RNA-DNA hybrids at a variety of locations throughout the genome. The R-loop, previously characterized in vitro at the leading strand replication origin (OH), is isolated as a native RNA-DNA hybrid copurifying with mtDNA. Surprisingly, other mitochondrial transcripts also form stable partial R-loops. These are abundant and affect mtDNA conformation. Current models regarding the mechanism of mammalian mtDNA replication have been expanded by recent data and discordant hypotheses. The presence of stable, nonreplicative, and partially hybridized RNA on the mtDNA template is significant for the reevaluation of replication models based on two-dimensional agarose gel analyses. In addition, the close association of a subpopulation of mtRNA with the DNA template has further implications regarding the structure, maintenance, and expression of the mitochondrial genome. These results demonstrate that variously processed and targeted mtRNAs within mammalian mitochondria likely have multiple functions in addition to their conventional roles.
Figures

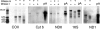
Similar articles
-
Mitochondrial DNA structure and expression in specialized subtypes of mammalian striated muscle.Mol Cell Biol. 1990 Nov;10(11):5671-8. doi: 10.1128/mcb.10.11.5671-5678.1990. Mol Cell Biol. 1990. PMID: 1700273 Free PMC article.
-
Replication of vertebrate mitochondrial DNA entails transient ribonucleotide incorporation throughout the lagging strand.EMBO J. 2006 Nov 15;25(22):5358-71. doi: 10.1038/sj.emboj.7601392. Epub 2006 Oct 26. EMBO J. 2006. PMID: 17066082 Free PMC article.
-
RNase mitochondrial RNA processing correctly cleaves a novel R loop at the mitochondrial DNA leading-strand origin of replication.Genes Dev. 1997 Mar 1;11(5):582-92. doi: 10.1101/gad.11.5.582. Genes Dev. 1997. PMID: 9119223
-
Transcription and replication of mitochondrial DNA.Hum Reprod. 2000 Jul;15 Suppl 2:11-7. doi: 10.1093/humrep/15.suppl_2.11. Hum Reprod. 2000. PMID: 11041509 Review.
-
An overview of mammalian mitochondrial DNA replication mechanisms.J Biochem. 2018 Sep 1;164(3):183-193. doi: 10.1093/jb/mvy058. J Biochem. 2018. PMID: 29931097 Free PMC article. Review.
Cited by
-
The structure of the mammalian RNase H2 complex provides insight into RNA.NA hybrid processing to prevent immune dysfunction.J Biol Chem. 2010 Feb 5;285(6):3617-3624. doi: 10.1074/jbc.M109.059048. Epub 2009 Nov 18. J Biol Chem. 2010. PMID: 19923215 Free PMC article.
-
Endonuclease G promotes hepatic mitochondrial respiration by selectively increasing mitochondrial tRNAThr production.Proc Natl Acad Sci U S A. 2025 Jan 7;122(1):e2411298122. doi: 10.1073/pnas.2411298122. Epub 2025 Jan 3. Proc Natl Acad Sci U S A. 2025. PMID: 39752519 Free PMC article.
-
Genome-wide distribution of RNA-DNA hybrids identifies RNase H targets in tRNA genes, retrotransposons and mitochondria.PLoS Genet. 2014 Oct 30;10(10):e1004716. doi: 10.1371/journal.pgen.1004716. eCollection 2014 Oct. PLoS Genet. 2014. PMID: 25357144 Free PMC article.
-
Length heterogeneity at conserved sequence block 2 in human mitochondrial DNA acts as a rheostat for RNA polymerase POLRMT activity.Nucleic Acids Res. 2016 Sep 19;44(16):7817-29. doi: 10.1093/nar/gkw648. Epub 2016 Jul 19. Nucleic Acids Res. 2016. PMID: 27436287 Free PMC article.
-
Mitochondrial DNA deletions are associated with non-B DNA conformations.Nucleic Acids Res. 2012 Sep;40(16):7606-21. doi: 10.1093/nar/gks500. Epub 2012 May 31. Nucleic Acids Res. 2012. PMID: 22661583 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

