Orientation-dependent interaction between Drosophila insulators is a property of this class of regulatory elements
- PMID: 18987002
- PMCID: PMC2602758
- DOI: 10.1093/nar/gkn781
Orientation-dependent interaction between Drosophila insulators is a property of this class of regulatory elements
Abstract
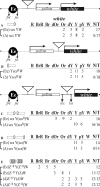
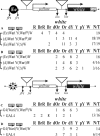
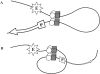
Insulators are defined as a class of regulatory elements that delimit independent transcriptional domains within eukaryotic genomes. According to previous data, an interaction (pairing) between some Drosophila insulators can support distant activation of a promoter by an enhancer. Here, we have demonstrated that pairs of well-studied insulators such as scs-scs, scs'-scs', 1A2-1A2 and Wari-Wari support distant activation of the white promoter by the yeast GAL4 activator in an orientation-dependent manner. The same is true for the efficiency of the enhancer that stimulates white expression in the eyes. In all insulator pairs tested, stimulation of the white gene was stronger when insulators were inserted between the eye enhancer or GAL4 and the white promoter in opposite orientations relative to each other. As shown previously, Zw5, Su(Hw) and dCTCF proteins are required for the functioning of different insulators that do not interact with each other. Here, strong functional interactions have been revealed between DNA fragments containing binding sites for either Zw5 or Su(Hw) or dCTCF protein but not between heterologous binding sites [Zw5-Su(Hw), dCTCF-Su(Hw), or dCTCF-Zw5]. These results suggest that insulator proteins can support selective interactions between distant regulatory elements.
Figures




Similar articles
-
New properties of Drosophila scs and scs' insulators.PLoS One. 2013 Apr 24;8(4):e62690. doi: 10.1371/journal.pone.0062690. Print 2013. PLoS One. 2013. PMID: 23638134 Free PMC article.
-
Red flag on the white reporter: a versatile insulator abuts the white gene in Drosophila and is omnipresent in mini-white constructs.Nucleic Acids Res. 2008 Feb;36(3):929-37. doi: 10.1093/nar/gkm992. Epub 2007 Dec 17. Nucleic Acids Res. 2008. PMID: 18086699 Free PMC article.
-
Insulators form gene loops by interacting with promoters in Drosophila.Development. 2011 Sep;138(18):4097-106. doi: 10.1242/dev.062836. Development. 2011. PMID: 21862564
-
[Roles of silencers from the regulatory region of the abdominal-B gene and the Zw5 and Su(Hw) insulators in regulating gene expression in Drosophila melanogaster].Genetika. 2004 Dec;40(12):1722-5. Genetika. 2004. PMID: 15648158 Review. Russian.
-
[The progress in the study of chromatin insulator].Yi Chuan. 2004 Jul;26(4):551-5. Yi Chuan. 2004. PMID: 15640060 Review. Chinese.
Cited by
-
Spermiogenesis and Male Fertility Require the Function of Suppressor of Hairy-Wing in Somatic Cyst Cells of Drosophila.Genetics. 2018 Jul;209(3):757-772. doi: 10.1534/genetics.118.301088. Epub 2018 May 8. Genetics. 2018. PMID: 29739818 Free PMC article.
-
The insulator protein SU(HW) fine-tunes nuclear lamina interactions of the Drosophila genome.PLoS One. 2010 Nov 24;5(11):e15013. doi: 10.1371/journal.pone.0015013. PLoS One. 2010. PMID: 21124834 Free PMC article.
-
Variants of the CNTNAP2 5' promoter as risk factors for autism spectrum disorders: a genetic and functional approach.Mol Psychiatry. 2015 Jul;20(7):839-49. doi: 10.1038/mp.2014.103. Epub 2014 Sep 16. Mol Psychiatry. 2015. PMID: 25224256
-
CTCF-binding elements mediate control of V(D)J recombination.Nature. 2011 Sep 11;477(7365):424-30. doi: 10.1038/nature10495. Nature. 2011. PMID: 21909113 Free PMC article.
-
Architectural proteins Pita, Zw5,and ZIPIC contain homodimerization domain and support specific long-range interactions in Drosophila.Nucleic Acids Res. 2016 Sep 6;44(15):7228-41. doi: 10.1093/nar/gkw371. Epub 2016 May 2. Nucleic Acids Res. 2016. PMID: 27137890 Free PMC article.
References
-
- Geyer PK, Corces VG. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev. 1992;6:1865–1873. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

