Mouse phenome database
- PMID: 18987003
- PMCID: PMC2686531
- DOI: 10.1093/nar/gkn778
Mouse phenome database
Abstract
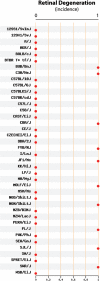
The Mouse Phenome Database (MPD; http://www.jax.org/phenome) is an open source, web-based repository of phenotypic and genotypic data on commonly used and genetically diverse inbred strains of mice and their derivatives. MPD is also a facility for query, analysis and in silico hypothesis testing. Currently MPD contains about 1400 phenotypic measurements contributed by research teams worldwide, including phenotypes relevant to human health such as cancer susceptibility, aging, obesity, susceptibility to infectious diseases, atherosclerosis, blood disorders and neurosensory disorders. Electronic access to centralized strain data enables investigators to select optimal strains for many systems-based research applications, including physiological studies, drug and toxicology testing, modeling disease processes and complex trait analysis. The ability to select strains for specific research applications by accessing existing phenotype data can bypass the need to (re)characterize strains, precluding major investments of time and resources. This functionality, in turn, accelerates research and leverages existing community resources. Since our last NAR reporting in 2007, MPD has added more community-contributed data covering more phenotypic domains and implemented several new tools and features, including a new interactive Tool Demo available through the MPD homepage (quick link: http://phenome.jax.org/phenome/trytools).
Figures






Similar articles
-
Mouse Phenome Database (MPD).Nucleic Acids Res. 2007 Jan;35(Database issue):D643-9. doi: 10.1093/nar/gkl1049. Epub 2006 Dec 6. Nucleic Acids Res. 2007. PMID: 17151079 Free PMC article.
-
Accessing Data Resources in the Mouse Phenome Database for Genetic Analysis of Murine Life Span and Health Span.J Gerontol A Biol Sci Med Sci. 2016 Feb;71(2):170-7. doi: 10.1093/gerona/glu223. Epub 2014 Dec 22. J Gerontol A Biol Sci Med Sci. 2016. PMID: 25533306 Free PMC article.
-
Mouse phenome database.Nucleic Acids Res. 2014 Jan;42(Database issue):D825-34. doi: 10.1093/nar/gkt1159. Epub 2013 Nov 15. Nucleic Acids Res. 2014. PMID: 24243846 Free PMC article.
-
Mouse phenome research: implications of genetic background.ILAR J. 2006;47(2):94-102. doi: 10.1093/ilar.47.2.94. ILAR J. 2006. PMID: 16547366 Review.
-
Bioinformatics resources for behavior studies in the laboratory mouse.Int Rev Neurobiol. 2012;104:71-90. doi: 10.1016/B978-0-12-398323-7.00004-5. Int Rev Neurobiol. 2012. PMID: 23195312 Review.
Cited by
-
Treatment- and population-dependent activity patterns of behavioral and expression QTLs.PLoS One. 2012;7(2):e31805. doi: 10.1371/journal.pone.0031805. Epub 2012 Feb 16. PLoS One. 2012. PMID: 22359631 Free PMC article.
-
Identification of a cardiac disease modifier gene using forward genetics in the mouse.PLoS Genet. 2009 Sep;5(9):e1000643. doi: 10.1371/journal.pgen.1000643. Epub 2009 Sep 18. PLoS Genet. 2009. PMID: 19763163 Free PMC article. No abstract available.
-
Transcriptome analysis of distinct mouse strains reveals kinesin light chain-1 splicing as an amyloid-β accumulation modifier.Proc Natl Acad Sci U S A. 2014 Feb 18;111(7):2638-43. doi: 10.1073/pnas.1307345111. Epub 2014 Feb 4. Proc Natl Acad Sci U S A. 2014. PMID: 24497505 Free PMC article.
-
Copy number variation influences gene expression and metabolic traits in mice.Hum Mol Genet. 2009 Nov 1;18(21):4118-29. doi: 10.1093/hmg/ddp360. Epub 2009 Jul 31. Hum Mol Genet. 2009. PMID: 19648292 Free PMC article.
-
Natural genetic variation profoundly regulates gene expression in immune cells and dictates susceptibility to CNS autoimmunity.Genes Immun. 2016 Dec;17(7):386-395. doi: 10.1038/gene.2016.37. Epub 2016 Sep 22. Genes Immun. 2016. PMID: 27653816 Free PMC article.
References
-
- Bogue M. Mouse Phenome Project: understanding human biology through mouse genetics and genomics. J. Appl. Physiol. 2003;95:1335–1337. - PubMed
-
- Grubb SC, Churchill GA, Bogue MA. A collaborative database of inbred mouse strain characteristics. Bioinformatics. 2004;20:2857–2859. - PubMed
-
- Mouse Phenotype Database Integration Consortium. Hancock JM, Adams NC, Aidinis V, Blake A, Bogue M, Brown SD, Chesler EJ, Davidson D, Duran C, Eppig JT, et al. Mouse Phenotype Database Integration Consortium: integration [corrected] of mouse phenome data resources. Mamm. Genome. 2007;18:157–163. (Errata in: Mamm. Genome. 2007; 18, 815. Mamm. Genome. 2008; 19, 219–220). - PMC - PubMed

