Reciprocal regulation of glycine-rich RNA-binding proteins via an interlocked feedback loop coupling alternative splicing to nonsense-mediated decay in Arabidopsis
- PMID: 18987006
- PMCID: PMC2602770
- DOI: 10.1093/nar/gkn847
Reciprocal regulation of glycine-rich RNA-binding proteins via an interlocked feedback loop coupling alternative splicing to nonsense-mediated decay in Arabidopsis
Abstract
The Arabidopsis RNA-binding protein AtGRP8 undergoes negative autoregulation at the post-transcriptional level. An elevated AtGRP8 protein level promotes the use of a cryptic 5' splice site to generate an alternatively spliced transcript, as_AtGRP8, retaining the 5' half of the intron with a premature termination codon. In mutants defective in nonsense-mediated decay (NMD) abundance of as_AtGRP8 but not its pre-mRNA is elevated, indicating that as_AtGRP8 is a direct NMD target, thus limiting the production of functional AtGRP8 protein. In addition to its own pre-mRNA, AtGRP8 negatively regulates the AtGRP7 transcript through promoting the formation of the equivalent alternatively spliced as_AtGRP7 transcript, leading to a decrease in AtGRP7 abundance. Recombinant AtGRP8 binds to its own and the AtGRP7 pre-mRNA, suggesting that this interaction is relevant for the splicing decision in vivo. AtGRP7 itself is part of a negative autoregulatory circuit that influences circadian oscillations of its own and the AtGRP8 transcript through alternative splicing linked to NMD. Thus, we identify an interlocked feedback loop through which two RNA-binding proteins autoregulate and reciprocally crossregulate by coupling unproductive splicing to NMD. A high degree of evolutionary sequence conservation in the introns retained in as_AtGRP8 or as_AtGRP7 points to an important function of these sequences.
Figures

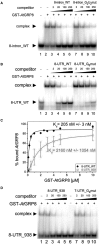
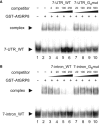





Similar articles
-
Auto-regulation of the circadian slave oscillator component AtGRP7 and regulation of its targets is impaired by a single RNA recognition motif point mutation.Plant J. 2007 Dec;52(6):1119-30. doi: 10.1111/j.1365-313X.2007.03302.x. Epub 2007 Oct 8. Plant J. 2007. PMID: 17924945
-
The circadian clock regulated RNA-binding protein AtGRP7 autoregulates its expression by influencing alternative splicing of its own pre-mRNA.Plant J. 2003 Jan;33(2):361-71. doi: 10.1046/j.1365-313x.2003.01629.x. Plant J. 2003. PMID: 12535349
-
A circadian clock-regulated toggle switch explains AtGRP7 and AtGRP8 oscillations in Arabidopsis thaliana.PLoS Comput Biol. 2013;9(3):e1002986. doi: 10.1371/journal.pcbi.1002986. Epub 2013 Mar 28. PLoS Comput Biol. 2013. PMID: 23555221 Free PMC article.
-
Plant serine/arginine-rich proteins: roles in precursor messenger RNA splicing, plant development, and stress responses.Wiley Interdiscip Rev RNA. 2011 Nov-Dec;2(6):875-89. doi: 10.1002/wrna.98. Epub 2011 Jul 15. Wiley Interdiscip Rev RNA. 2011. PMID: 21766458 Review.
-
Role of plant RNA-binding proteins in development, stress response and genome organization.Trends Plant Sci. 2009 Apr;14(4):229-36. doi: 10.1016/j.tplants.2009.01.007. Epub 2009 Mar 13. Trends Plant Sci. 2009. PMID: 19285908 Review.
Cited by
-
Beyond Transcription: Fine-Tuning of Circadian Timekeeping by Post-Transcriptional Regulation.Genes (Basel). 2018 Dec 10;9(12):616. doi: 10.3390/genes9120616. Genes (Basel). 2018. PMID: 30544736 Free PMC article. Review.
-
The Plant Circadian Oscillator.Biology (Basel). 2019 Mar 12;8(1):14. doi: 10.3390/biology8010014. Biology (Basel). 2019. PMID: 30870980 Free PMC article. Review.
-
Evolutionarily Conserved Alternative Splicing Across Monocots.Genetics. 2017 Oct;207(2):465-480. doi: 10.1534/genetics.117.300189. Epub 2017 Aug 24. Genetics. 2017. PMID: 28839042 Free PMC article.
-
Network news: prime time for systems biology of the plant circadian clock.Curr Opin Genet Dev. 2010 Dec;20(6):588-98. doi: 10.1016/j.gde.2010.08.010. Curr Opin Genet Dev. 2010. PMID: 20889330 Free PMC article. Review.
-
Alternative splicing in plants--coming of age.Trends Plant Sci. 2012 Oct;17(10):616-23. doi: 10.1016/j.tplants.2012.06.001. Epub 2012 Jun 27. Trends Plant Sci. 2012. PMID: 22743067 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

