The GTOP database in 2009: updated content and novel features to expand and deepen insights into protein structures and functions
- PMID: 18987007
- PMCID: PMC2686575
- DOI: 10.1093/nar/gkn855
The GTOP database in 2009: updated content and novel features to expand and deepen insights into protein structures and functions
Abstract
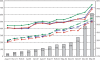
The Genomes TO Protein Structures and Functions (GTOP) database (http://spock.genes.nig.ac.jp/~genome/gtop.html) freely provides an extensive collection of information on protein structures and functions obtained by application of various computational tools to the amino acid sequences of entirely sequenced genomes. GTOP contains annotations of 3D structures, protein families, functions, and other useful data of a protein of interest in user-friendly ways to give a deep insight into the protein structure. From the initial 1999 version, GTOP has been continually updated to reap the fruits of genome projects and augmented to supply novel information, in particular intrinsically disordered regions. As intrinsically disordered regions constitute a considerable fraction of proteins and often play crucial roles especially in eukaryotes, their assignments give important additional clues to the functionality of proteins. Additionally, we have incorporated the following features into GTOP: a platform independent structural viewer, results of HMM searches against SCOP and Pfam, secondary structure predictions, color display of exon boundaries in eukaryotic proteins, assignments of gene ontology terms, search tools, and master files.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

