BloodExpress: a database of gene expression in mouse haematopoiesis
- PMID: 18987008
- PMCID: PMC2686428
- DOI: 10.1093/nar/gkn854
BloodExpress: a database of gene expression in mouse haematopoiesis
Abstract
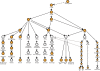
Haematopoiesis is the process whereby blood stem cells give rise to at least fourteen functionally distinct mature cell types, and represents the best characterized mammalian adult stem cell system. Here we introduce the BloodExpress database, the first public resource integrating mouse blood cell expression profiles. BloodExpress enables the searching of data from individual studies in a single database accessible through a user-friendly web interface. Microarray datasets have been processed uniformly to allow their comparison on the BloodExpress platform. BloodExpress covers the majority of murine blood cell types, including both progenitors and terminally differentiated cells. This allows for the identification of dynamic changes in gene expression as cells differentiate down the well-defined haematopoietic hierarchy. A gene-centric interface returns haematopoietic expression patterns together with functional annotation and a list of other genes with similar expression patterns. A cell type-centric interface allows the identification of genes expressed at specific points of blood development, with the additional and useful capability of filtering by specific gene functional categories. BloodExpress thus constitutes a platform for the discovery of novel gene functions across the haematopoietic tree. BloodExpress is freely accessible at http://hscl.cimr.cam.ac.uk/bloodexpress/.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

