The Arabidopsis A4 subfamily of lectin receptor kinases negatively regulates abscisic acid response in seed germination
- PMID: 18987212
- PMCID: PMC2613733
- DOI: 10.1104/pp.108.130583
The Arabidopsis A4 subfamily of lectin receptor kinases negatively regulates abscisic acid response in seed germination
Abstract
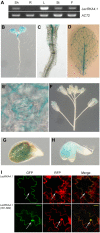
Abscisic acid (ABA) is an important plant hormone for a wide array of growth and developmental processes and stress responses, but the mechanism of ABA signal perception on the plasma membrane remains to be dissected. A previous GeneChip analysis revealed that a member of the A4 subfamily of lectin receptor kinases (LecRKs) of Arabidopsis (Arabidopsis thaliana), At5g01540 (designated LecRKA4.1), is up-regulated in response to a low dose of ABA in the rop10-1 background. Here, we present functional evidence to support its role in ABA response. LecRKA4.1 is expressed in seeds and leaves but not in roots, and the protein is localized to the plasma membrane. A T-DNA knockout mutant, lecrka4.1-1, slightly enhanced ABA inhibition of seed germination. Interestingly, LecRKA4.1 is adjacent to two other members of the A4 subfamily of LecRK genes, At5g01550 (LecRKA4.2) and At5g01560 (LecRKA4.3). We found that loss-of-function mutants of LecRKA4.2 and LecRKA4.3 exhibited similarly weak enhancement of ABA response in seed germination inhibition. Furthermore, LecRKA4.2 suppression by RNA interference in lecrka4.1-1 showed stronger ABA inhibition of seed germination than lecrka4.1-1, while the response to gibberellic acid was not affected in lecrka4.1-1 and lecrka4.1-1; LecRKA4.2 (RNAi) lines. Expression studies, together with network-based analysis, suggest that LecRKA4.1 and LecRKA4.2 regulate some of the ABA-responsive genes. Taken together, our results demonstrate that the A4 subfamily of LecRKs has a redundant function in the negative regulation of ABA response in seed germination.
Figures




Similar articles
-
Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy.Plant Cell Physiol. 2009 Jul;50(7):1345-63. doi: 10.1093/pcp/pcp083. Epub 2009 Jun 18. Plant Cell Physiol. 2009. PMID: 19541597
-
Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis.Plant Cell. 2007 Feb;19(2):485-94. doi: 10.1105/tpc.106.048538. Epub 2007 Feb 16. Plant Cell. 2007. PMID: 17307925 Free PMC article.
-
The Arabidopsis tandem zinc finger protein AtTZF1 affects ABA- and GA-mediated growth, stress and gene expression responses.Plant J. 2011 Jan;65(2):253-68. doi: 10.1111/j.1365-313X.2010.04419.x. Epub 2010 Dec 8. Plant J. 2011. PMID: 21223390
-
Diverse functional interactions between nitric oxide and abscisic acid in plant development and responses to stress.J Exp Bot. 2014 Mar;65(4):907-21. doi: 10.1093/jxb/ert454. Epub 2013 Dec 26. J Exp Bot. 2014. PMID: 24371253 Review.
-
[Abscisic acid metabolism].Postepy Biochem. 2013;59(1):83-8. Postepy Biochem. 2013. PMID: 23821946 Review. Polish.
Cited by
-
The receptor kinase IMPAIRED OOMYCETE SUSCEPTIBILITY1 attenuates abscisic acid responses in Arabidopsis.Plant Physiol. 2014 Nov;166(3):1506-18. doi: 10.1104/pp.114.248518. Epub 2014 Oct 1. Plant Physiol. 2014. PMID: 25274985 Free PMC article.
-
Prediction and validation of cis-regulatory elements in 5' upstream regulatory regions of lectin receptor-like kinase gene family in rice.Protoplasma. 2017 Mar;254(2):669-684. doi: 10.1007/s00709-016-0979-6. Epub 2016 May 18. Protoplasma. 2017. PMID: 27193099
-
Lectin receptor kinases in plant innate immunity.Front Plant Sci. 2013 May 7;4:124. doi: 10.3389/fpls.2013.00124. eCollection 2013. Front Plant Sci. 2013. PMID: 23675375 Free PMC article.
-
Genome-wide identification and functional exploration of the legume lectin genes in Brassica napus and their roles in Sclerotinia disease resistance.Front Plant Sci. 2022 Jul 22;13:963263. doi: 10.3389/fpls.2022.963263. eCollection 2022. Front Plant Sci. 2022. PMID: 35968144 Free PMC article.
-
Combining population genomics and fitness QTLs to identify the genetics of local adaptation in Arabidopsis thaliana.Proc Natl Acad Sci U S A. 2018 May 8;115(19):5028-5033. doi: 10.1073/pnas.1719998115. Epub 2018 Apr 23. Proc Natl Acad Sci U S A. 2018. PMID: 29686078 Free PMC article.
References
-
- Andre S, Siebert HC, Nishiguchi M, Tazaki K, Gabius HJ (2005) Evidence for lectin activity of a plant receptor-like protein kinase by application of neoglycoproteins and bioinformatic algorithms. Biochim Biophys Acta 1725 222–232 - PubMed
-
- Barre A, Herve C, Lescure B, Rouge P (2002) Lectin receptor kinases in plants. Crit Rev Plant Sci 21 379–399
-
- Chen X, Shang J, Chen D, Lei C, Zou Y, Zhai W, Liu G, Xu J, Ling Z, Cao G, et al (2006) A B-lectin receptor kinase gene conferring rice blast resistance. Plant J 46 794–804 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

