GRASSIUS: a platform for comparative regulatory genomics across the grasses
- PMID: 18987217
- PMCID: PMC2613736
- DOI: 10.1104/pp.108.128579
GRASSIUS: a platform for comparative regulatory genomics across the grasses
Abstract
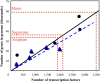
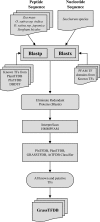
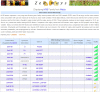
Transcription factors (TFs) are major players in gene regulatory networks and interactions between TFs and their target genes furnish spatiotemporal patterns of gene expression. Establishing the architecture of regulatory networks requires gathering information on TFs, their targets in the genome, and the corresponding binding sites. We have developed GRASSIUS (Grass Regulatory Information Services) as a knowledge-based Web resource that integrates information on TFs and gene promoters across the grasses. In its initial implementation, GRASSIUS consists of two separate, yet linked, databases. GrassTFDB holds information on TFs from maize (Zea mays), sorghum (Sorghum bicolor), sugarcane (Saccharum spp.), and rice (Oryza sativa). TFs are classified into families and phylogenetic relationships begin to uncover orthologous relationships among the participating species. This database also provides a centralized clearinghouse for TF synonyms in the grasses. GrassTFDB is linked to the grass TFome collection, which provides clones in recombination-based vectors corresponding to full-length open reading frames for a growing number of grass TFs. GrassPROMDB contains promoter and cis-regulatory element information for those grass species and genes for which enough data are available. The integration of GrassTFDB and GrassPROMDB will be accomplished through GrassRegNet as a first step in representing the architecture of grass regulatory networks. GRASSIUS can be accessed from www.grassius.org.
Figures



References
-
- Babu MM, Luscombe NM, Aravind L, Gerstein M, Teichmann SA (2004) Structure and evolution of transcriptional regulatory networks. Curr Opin Struct Biol 14 283–291 - PubMed
-
- Bennetzen JL (2007) Patterns in grass genome evolution. Curr Opin Plant Biol 10 176–181 - PubMed
-
- Braun EL, Grotewold E (2001) Fungal Zuotin proteins evolved from MIDA1-like factors by lineage-specific loss of MYB domains. Mol Biol Evol 18 1401–1412 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

