Metagenome analysis of an extreme microbial symbiosis reveals eurythermal adaptation and metabolic flexibility
- PMID: 18987310
- PMCID: PMC2579889
- DOI: 10.1073/pnas.0802782105
Metagenome analysis of an extreme microbial symbiosis reveals eurythermal adaptation and metabolic flexibility
Abstract
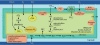
Hydrothermal vent ecosystems support diverse life forms, many of which rely on symbiotic associations to perform functions integral to survival in these extreme physicochemical environments. Epsilonproteobacteria, found free-living and in intimate associations with vent invertebrates, are the predominant vent-associated microorganisms. The vent-associated polychaete worm, Alvinella pompejana, is host to a visibly dense fleece of episymbionts on its dorsal surface. The episymbionts are a multispecies consortium of Epsilonproteobacteria present as a biofilm. We unraveled details of these enigmatic, uncultivated episymbionts using environmental genome sequencing. They harbor wide-ranging adaptive traits that include high levels of strain variability analogous to Epsilonproteobacteria pathogens such as Helicobacter pylori, metabolic diversity of free-living bacteria, and numerous orthologs of proteins that we hypothesize are each optimally adapted to specific temperature ranges within the 10-65 degrees C fluctuations characteristic of the A. pompejana habitat. This strategic combination enables the consortium to thrive under diverse thermal and chemical regimes. The episymbionts are metabolically tuned for growth in hydrothermal vent ecosystems with genes encoding the complete rTCA cycle, sulfur oxidation, and denitrification; in addition, the episymbiont metagenome also encodes capacity for heterotrophic and aerobic metabolisms. Analysis of the environmental genome suggests that A. pompejana may benefit from the episymbionts serving as a stable source of food and vitamins. The success of Epsilonproteobacteria as episymbionts in hydrothermal vent ecosystems is a product of adaptive capabilities, broad metabolic capacity, strain variance, and virulent traits in common with pathogens.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Hurtado LA, Lutz RA, Vrijenhoek RC. Distinct patterns of genetic differentiation among annelids of eastern Pacific hydrothermal vents. Mol Ecol. 2004;13:2603–2615. - PubMed
-
- Le Bris N, Zbinden M, Gaill F. Processes controlling the physico-chemical micro-environments associated with Pompeii worms. Deep-Sea Res I. 2005;52:1071–1083.
-
- Di Meo-Savoie CA, Luther GW, Cary SC. Physicochemical characterization of the microhabitat of the epibionts associated with Alvinella pompejana, a hydrothermal vent annelid. Geochim Cosmochim Acta. 2004;68:2055–2066.
-
- Luther GW, et al. Chemical speciation drives hydrothermal vent ecology. Nature. 2001;410:813–816. - PubMed
-
- Gaill F, Mann K, Wiedemann H, Engel J, Timpl R. Structural comparison of cuticle and interstitial collagens from annelids living in shallow sea-water and at deep-sea hydrothermal vents. J Mol Biol. 1995;246:284–294. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

