Alternative requirements for Vestigial, Scalloped, and Dmef2 during muscle differentiation in Drosophila melanogaster
- PMID: 18987343
- PMCID: PMC2613084
- DOI: 10.1091/mbc.e08-03-0288
Alternative requirements for Vestigial, Scalloped, and Dmef2 during muscle differentiation in Drosophila melanogaster
Abstract
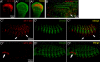
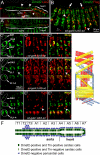
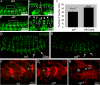
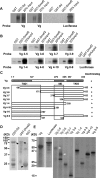
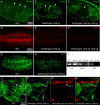
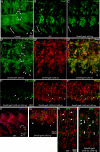
Vertebrate development requires the activity of the myocyte enhancer factor 2 (mef2) gene family for muscle cell specification and subsequent differentiation. Additionally, several muscle-specific functions of MEF2 family proteins require binding additional cofactors including members of the Transcription Enhancing Factor-1 (TEF-1) and Vestigial-like protein families. In Drosophila there is a single mef2 (Dmef2) gene as well single homologues of TEF-1 and vestigial-like, scalloped (sd), and vestigial (vg), respectively. To clarify the role(s) of these factors, we examined the requirements for Vg and Sd during Drosophila muscle specification. We found that both are required for muscle differentiation as loss of sd or vg leads to a reproducible loss of a subset of either cardiac or somatic muscle cells in developing embryos. This muscle requirement for Sd or Vg is cell specific, as ubiquitous overexpression of either or both of these proteins in muscle cells has a deleterious effect on muscle differentiation. Finally, using both in vitro and in vivo binding assays, we determined that Sd, Vg, and Dmef2 can interact directly. Thus, the muscle-specific phenotypes we have associated with Vg or Sd may be a consequence of alternative binding of Vg and/or Sd to Dmef2 forming alternative protein complexes that modify Dmef2 activity.
Figures


References
-
- Azpiazu N., Frasch M. tinman and bagpipe: two homeo box genes that determine cell fates in the dorsal mesoderm of Drosophila. Genes Dev. 1993;7:1325–1340. - PubMed
-
- Bao M. Z., Schwartz M. A., Cantin G. T., Yates J. R., 3rd, Madhani H. D. Pheromone-dependent destruction of the Tec1 transcription factor is required for MAP kinase signaling specificity in yeast. Cell. 2004;119:991–1000. - PubMed
-
- Bate M., Rushton E. Myogenesis and muscle patterning in Drosophila. C. R. Acad. Sci. III. 1993;316:1047–1061. - PubMed
-
- Baylies M. K., Bate M., Ruiz Gomez M. Myogenesis: a view from Drosophila. Cell. 1998;93:921–927. - PubMed
-
- Bernard F., Lalouette A., Gullaud M., Jeantet A. Y., Cossard R., Zider A., Ferveur J. F., Silber J. Control of apterous by vestigial drives indirect flight muscle development in Drosophila. Dev. Biol. 2003;260:391–403. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

