The transcription factor c-Myc enhances KIR gene transcription through direct binding to an upstream distal promoter element
- PMID: 18987359
- PMCID: PMC2665893
- DOI: 10.1182/blood-2008-07-166389
The transcription factor c-Myc enhances KIR gene transcription through direct binding to an upstream distal promoter element
Abstract
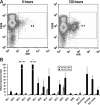
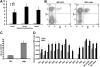
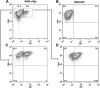
The killer cell immunoglobulin-like receptor (KIR) repertoire of natural killer (NK) cells determines their ability to detect infected or transformed target cells. Although epigenetic mechanisms play a role in KIR gene expression, work in the mouse suggests that other regulatory elements may be involved at specific stages of NK-cell development. Here we report the effects of the transcription factor c-Myc on KIR expression. c-Myc directly binds to, and promotes transcription from, a distal element identified upstream of most KIR genes. Binding of endogenous c-Myc to the distal promoter element is significantly enhanced upon interleukin-15 (IL-15) stimulation in peripheral blood NK cells and correlates with an increase in KIR transcription. In addition, the overexpression of c-Myc during NK-cell development promotes transcription from the distal promoter element and contributes to the overall transcription of multiple KIR genes. Our data demonstrate the significance of the 5' promoter element upstream of the conventional KIR promoter region and support a model whereby IL-15 stimulates c-Myc binding at the distal KIR promoter during NK-cell development to promote KIR transcription. This finding provides a direct link between NK-cell activation signals and KIR expression required for acquisition of effector function during NK-cell education.
Figures




References
-
- Katz G, Markel G, Mizrahi S, Arnon TI, Mandelboim O. Recognition of HLA-Cw4 but not HLA-Cw6 by the NK cell receptor killer cell Ig-like receptor two-domain short tail number 4. J Immunol. 2001;166:7260–7267. - PubMed
-
- Valiante N, Uhberg M, Shilling H, et al. Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity. 1997;7:739–751. - PubMed
-
- Terme M, Ullrich E, Delahaye NF, Chaput N, Zitvogel L. Natural killer cell-directed therapies: moving from unexpected results to successful strategies. Nat Immunol. 2008;9:486–494. - PubMed
-
- Khakoo SI, Thio CL, Martin MP, et al. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305:872–874. - PubMed
-
- Martin MP, Gao X, Lee JH, et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet. 2002;31:429–434. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

