X-ray structure of NS1 from a highly pathogenic H5N1 influenza virus
- PMID: 18987632
- PMCID: PMC2798118
- DOI: 10.1038/nature07444
X-ray structure of NS1 from a highly pathogenic H5N1 influenza virus
Abstract
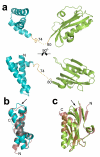
The recent emergence of highly pathogenic avian (H5N1) influenza viruses, their epizootic and panzootic nature, and their association with lethal human infections have raised significant global health concerns. Several studies have underlined the importance of non-structural protein NS1 in the increased pathogenicity and virulence of these strains. NS1, which consists of two domains-a double-stranded RNA (dsRNA) binding domain and the effector domain, separated through a linker-is an antagonist of antiviral type-I interferon response in the host. Here we report the X-ray structure of the full-length NS1 from an H5N1 strain (A/Vietnam/1203/2004) that was associated with 60% of human deaths in an outbreak in Vietnam. Compared to the individually determined structures of the RNA binding domain and the effector domain from non-H5N1 strains, the RNA binding domain within H5N1 NS1 exhibits modest structural changes, while the H5N1 effector domain shows significant alteration, particularly in the dimeric interface. Although both domains in the full-length NS1 individually participate in dimeric interactions, an unexpected finding is that these interactions result in the formation of a chain of NS1 molecules instead of distinct dimeric units. Three such chains in the crystal interact with one another extensively to form a tubular organization of similar dimensions to that observed in the cryo-electron microscopy images of NS1 in the presence of dsRNA. The tubular oligomeric organization of NS1, in which residues implicated in dsRNA binding face a 20-A-wide central tunnel, provides a plausible mechanism for how NS1 sequesters varying lengths of dsRNA, to counter cellular antiviral dsRNA response pathways, while simultaneously interacting with other cellular ligands during an infection.
Figures


References
-
- Abdel-Ghafar AN, et al. Update on avian influenza A (H5N1) virus infection in humans. N Engl J Med. 2008;358:261–273. - PubMed
-
- Beigel JH, et al. Avian influenza A (H5N1) infection in humans. N Engl J Med. 2005;353:1374–1385. - PubMed
-
- Seo SH, Hoffmann E, Webster RG. The NS1 gene of H5N1 influenza viruses circumvents the host anti-viral cytokine responses. Virus Res. 2004;103:107–113. - PubMed
-
- Seo SH, Hoffmann E, Webster RG. Lethal H5N1 influenza viruses escape host anti-viral cytokine responses. Nat Med. 2002;8:950–954. - PubMed
-
- Chien CY, et al. A novel RNA-binding motif in influenza A virus non-structural protein 1. Nat Struct Biol. 1997;4:891–895. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

