Visualizing the dynamics of viral replication in living cells via Tat peptide delivery of nuclease-resistant molecular beacons
- PMID: 18988730
- PMCID: PMC2582299
- DOI: 10.1073/pnas.0807066105
Visualizing the dynamics of viral replication in living cells via Tat peptide delivery of nuclease-resistant molecular beacons
Abstract
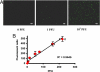
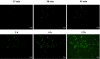
In this study, we describe the use of nuclease-resistant molecular beacons (MBs) for the real-time detection of coxsackievirus B6 replication in living Buffalo green monkey kidney (BGMK) cells via Tat peptide delivery. A nuclease-resistant MB containing 2'-O-methyl RNA bases with phosphorothioate internucleotide linkages was designed to specifically target an 18-bp 5' noncoding region of the viral genome. For intracellular delivery, a cell-penetrating Tat peptide was conjugated to the MB by using a thiol-maleimide linkage. Presence of the Tat peptide enabled nearly 100% intracellular delivery within 15 min. When the conjugate was introduced into BGMK cell monolayers infected with coxsackievirus B6, a discernible fluorescence was observed at 30 min after infection, and as few as 1 infectious viral particle could be detected within 2 h. The stability and the intracellular delivery properties of the modified MBs enabled real-time monitoring of the cell-to-cell spreading of viral infection. These results suggest that the Tat-modified, nuclease-resistant MBs may be powerful tools for improving our understanding of the dynamic behavior of viral replication and for therapeutic studies of antiviral treatments.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Comment in
-
Shedding light on virus replication.Proc Natl Acad Sci U S A. 2008 Nov 11;105(45):17213-4. doi: 10.1073/pnas.0809841105. Epub 2008 Nov 7. Proc Natl Acad Sci U S A. 2008. PMID: 18997007 Free PMC article. No abstract available.
Similar articles
-
Visualization and detection of infectious coxsackievirus replication using a combined cell culture-molecular beacon assay.Appl Environ Microbiol. 2005 Dec;71(12):8397-401. doi: 10.1128/AEM.71.12.8397-8401.2005. Appl Environ Microbiol. 2005. PMID: 16332827 Free PMC article.
-
Shedding light on virus replication.Proc Natl Acad Sci U S A. 2008 Nov 11;105(45):17213-4. doi: 10.1073/pnas.0809841105. Epub 2008 Nov 7. Proc Natl Acad Sci U S A. 2008. PMID: 18997007 Free PMC article. No abstract available.
-
Molecular beacon-quantum dot-Au nanoparticle hybrid nanoprobes for visualizing virus replication in living cells.Chem Commun (Camb). 2010 Jun 14;46(22):3914-6. doi: 10.1039/c001553a. Epub 2010 Apr 23. Chem Commun (Camb). 2010. PMID: 20419176
-
Infection and replication of Tat- human immunodeficiency viruses: genetic analyses of LTR and tat mutations in primary and long-term human lymphoid cells.Virology. 1995 Aug 1;211(1):157-69. doi: 10.1006/viro.1995.1388. Virology. 1995. PMID: 7645208
-
RNA recognition and regulation of HIV-1 gene expression by viral factor Tat.Biochemistry (Mosc). 1998 May;63(5):489-503. Biochemistry (Mosc). 1998. PMID: 9632883 Review.
Cited by
-
Optimizing peptide nucleic acid probes for hybridization-based detection and identification of bacterial pathogens.Analyst. 2019 Feb 25;144(5):1565-1574. doi: 10.1039/c8an02194e. Analyst. 2019. PMID: 30656297 Free PMC article.
-
Engineering Novel Molecular Beacon Constructs to Study Intracellular RNA Dynamics and Localization.Genomics Proteomics Bioinformatics. 2017 Oct;15(5):279-286. doi: 10.1016/j.gpb.2017.04.004. Epub 2017 Sep 21. Genomics Proteomics Bioinformatics. 2017. PMID: 28942262 Free PMC article. Review.
-
A molecular beacon-based approach for live-cell imaging of RNA transcripts with minimal target engineering at the single-molecule level.Sci Rep. 2017 May 8;7(1):1550. doi: 10.1038/s41598-017-01740-1. Sci Rep. 2017. PMID: 28484218 Free PMC article.
-
Imaging intracellular RNA distribution and dynamics in living cells.Nat Methods. 2009 May;6(5):331-8. doi: 10.1038/nmeth.1321. Nat Methods. 2009. PMID: 19404252 Review.
-
Real-time imaging of the epithelial-mesenchymal transition using microRNA-200a sequence-based molecular beacon-conjugated magnetic nanoparticles.PLoS One. 2014 Jul 21;9(7):e102164. doi: 10.1371/journal.pone.0102164. eCollection 2014. PLoS One. 2014. PMID: 25048580 Free PMC article.
References
-
- Tyagi S, Kramer FR. Molecular beacons: Probes that fluoresce upon hybridization. Nat Biotechnol. 1996;14:303–308. - PubMed
-
- Marras SA, Kramer FR, Tyagi S. Multiplex detection of single-nucleotide variations using molecular beacons. Genet Anal. 1999;14:151–156. - PubMed
-
- Tyagi S, Bratu DP, Kramer FR. Multicolor molecular beacons for allele discrimination. Nat Biotechnol. 1997;16:49–53. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

