Genetic mapping in human disease
- PMID: 18988837
- PMCID: PMC2694957
- DOI: 10.1126/science.1156409
Genetic mapping in human disease
Abstract
Genetic mapping provides a powerful approach to identify genes and biological processes underlying any trait influenced by inheritance, including human diseases. We discuss the intellectual foundations of genetic mapping of Mendelian and complex traits in humans, examine lessons emerging from linkage analysis of Mendelian diseases and genome-wide association studies of common diseases, and discuss questions and challenges that lie ahead.
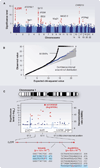
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

