Insights into translational termination from the structure of RF2 bound to the ribosome
- PMID: 18988853
- PMCID: PMC2642913
- DOI: 10.1126/science.1164840
Insights into translational termination from the structure of RF2 bound to the ribosome
Abstract
The termination of protein synthesis occurs through the specific recognition of a stop codon in the A site of the ribosome by a release factor (RF), which then catalyzes the hydrolysis of the nascent protein chain from the P-site transfer RNA. Here we present, at a resolution of 3.5 angstroms, the crystal structure of RF2 in complex with its cognate UGA stop codon in the 70S ribosome. The structure provides insight into how RF2 specifically recognizes the stop codon; it also suggests a model for the role of a universally conserved GGQ motif in the catalysis of peptide release.
Figures

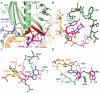
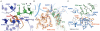
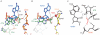
Comment in
-
Biochemistry. Getting close to termination.Science. 2008 Nov 7;322(5903):863-5. doi: 10.1126/science.1166913. Science. 2008. PMID: 18988828 No abstract available.
References
-
- Youngman EM, McDonald ME, Green R. Annu Rev Microbiol. 2008;62:353. - PubMed
-
-
Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.
-
-
- Ito K, Uno M, Nakamura Y. Nature. 2000;403:680. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

