The transcriptional repressor protein NsrR senses nitric oxide directly via a [2Fe-2S] cluster
- PMID: 18989365
- PMCID: PMC2577008
- DOI: 10.1371/journal.pone.0003623
The transcriptional repressor protein NsrR senses nitric oxide directly via a [2Fe-2S] cluster
Abstract
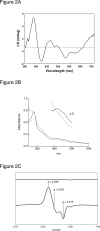
The regulatory protein NsrR, a member of the Rrf2 family of transcription repressors, is specifically dedicated to sensing nitric oxide (NO) in a variety of pathogenic and non-pathogenic bacteria. It has been proposed that NO directly modulates NsrR activity by interacting with a predicted [Fe-S] cluster in the NsrR protein, but no experimental evidence has been published to support this hypothesis. Here we report the purification of NsrR from the obligate aerobe Streptomyces coelicolor. We demonstrate using UV-visible, near UV CD and EPR spectroscopy that the protein contains an NO-sensitive [2Fe-2S] cluster when purified from E. coli. Upon exposure of NsrR to NO, the cluster is nitrosylated, which results in the loss of DNA binding activity as detected by bandshift assays. Removal of the [2Fe-2S] cluster to generate apo-NsrR also resulted in loss of DNA binding activity. This is the first demonstration that NsrR contains an NO-sensitive [2Fe-2S] cluster that is required for DNA binding activity.
Conflict of interest statement
Figures


References
-
- Lane N. Climate change: What's in the rising tide? Nature. 2007;449:778–780. - PubMed
-
- D'Autreaux B, Tucker NP, Dixon R, Spiro S. A non-haem iron centre in the transcription factor NorR senses nitric oxide. Nature. 2005;437:769–772. - PubMed
-
- Gardner AM, Helmick RA, Gardner PR. Flavorubredoxin, an inducible catalyst for nitric oxide reduction and detoxification in Escherichia coli. J Biol Chem. 2002;277:8172–8177. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BB/F009429/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/D009588/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- A51065X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/J/00000016/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- F009429/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

