Transmembrane topology and signal peptide prediction using dynamic bayesian networks
- PMID: 18989393
- PMCID: PMC2570248
- DOI: 10.1371/journal.pcbi.1000213
Transmembrane topology and signal peptide prediction using dynamic bayesian networks
Abstract
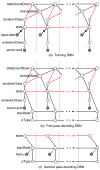
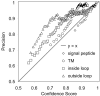
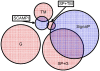
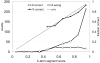
Hidden Markov models (HMMs) have been successfully applied to the tasks of transmembrane protein topology prediction and signal peptide prediction. In this paper we expand upon this work by making use of the more powerful class of dynamic Bayesian networks (DBNs). Our model, Philius, is inspired by a previously published HMM, Phobius, and combines a signal peptide submodel with a transmembrane submodel. We introduce a two-stage DBN decoder that combines the power of posterior decoding with the grammar constraints of Viterbi-style decoding. Philius also provides protein type, segment, and topology confidence metrics to aid in the interpretation of the predictions. We report a relative improvement of 13% over Phobius in full-topology prediction accuracy on transmembrane proteins, and a sensitivity and specificity of 0.96 in detecting signal peptides. We also show that our confidence metrics correlate well with the observed precision. In addition, we have made predictions on all 6.3 million proteins in the Yeast Resource Center (YRC) database. This large-scale study provides an overall picture of the relative numbers of proteins that include a signal-peptide and/or one or more transmembrane segments as well as a valuable resource for the scientific community. All DBNs are implemented using the Graphical Models Toolkit. Source code for the models described here is available at http://noble.gs.washington.edu/proj/philius. A Philius Web server is available at http://www.yeastrc.org/philius, and the predictions on the YRC database are available at http://www.yeastrc.org/pdr.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


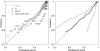

References
-
- Sonnhammer E, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc ISMB. 1998;6:175–182. - PubMed
-
- Tusnady G, Simon I. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J Mol Biol. 1998;283:489–506. - PubMed
-
- Schwartz R, Chow YL. The N-best algorithms: an efficient and exact procedure for finding the N most likely sentence hypotheses. IEEE Int Conf Acoust Speech Signal Process. 1990;1:81–84.
-
- Tusnady G, Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics. 2001;17:849–850. - PubMed
-
- Nielsen H, Krogh A. Prediction of signal peptides and signal anchors by a hidden Markov model. Proc ISMB. 1998;6:122–30. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

