Inferring pathway activity toward precise disease classification
- PMID: 18989396
- PMCID: PMC2563693
- DOI: 10.1371/journal.pcbi.1000217
Inferring pathway activity toward precise disease classification
Abstract
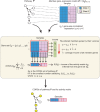
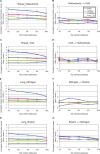
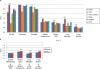
The advent of microarray technology has made it possible to classify disease states based on gene expression profiles of patients. Typically, marker genes are selected by measuring the power of their expression profiles to discriminate among patients of different disease states. However, expression-based classification can be challenging in complex diseases due to factors such as cellular heterogeneity within a tissue sample and genetic heterogeneity across patients. A promising technique for coping with these challenges is to incorporate pathway information into the disease classification procedure in order to classify disease based on the activity of entire signaling pathways or protein complexes rather than on the expression levels of individual genes or proteins. We propose a new classification method based on pathway activities inferred for each patient. For each pathway, an activity level is summarized from the gene expression levels of its condition-responsive genes (CORGs), defined as the subset of genes in the pathway whose combined expression delivers optimal discriminative power for the disease phenotype. We show that classifiers using pathway activity achieve better performance than classifiers based on individual gene expression, for both simple and complex case-control studies including differentiation of perturbed from non-perturbed cells and subtyping of several different kinds of cancer. Moreover, the new method outperforms several previous approaches that use a static (i.e., non-conditional) definition of pathways. Within a pathway, the identified CORGs may facilitate the development of better diagnostic markers and the discovery of core alterations in human disease.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. - PubMed
-
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. - PubMed
-
- Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33:49–54. - PubMed
-
- van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. - PubMed
-
- Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–679. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

