Mechanistic insight into the ribosome biogenesis functions of the ancient protein KsgA
- PMID: 18990185
- PMCID: PMC2709978
- DOI: 10.1111/j.1365-2958.2008.06485.x
Mechanistic insight into the ribosome biogenesis functions of the ancient protein KsgA
Abstract
While the general blueprint of ribosome biogenesis is evolutionarily conserved, most details have diverged considerably. A striking exception to this divergence is the universally conserved KsgA/Dim1p enzyme family, which modifies two adjacent adenosines in the terminal helix of small subunit ribosomal RNA (rRNA). While localization of KsgA on 30S subunits [small ribosomal subunits (SSUs)] and genetic interaction data have suggested that KsgA acts as a ribosome biogenesis factor, mechanistic details and a rationale for its extreme conservation are still lacking. To begin to address these questions we have characterized the function of Escherichia coli KsgA in vivo using both a ksgA deletion strain and a methyltransferase-deficient form of this protein. Our data reveal cold sensitivity and altered ribosomal profiles are associated with a DeltaksgA genotype in E. coli. Our work also indicates that loss of KsgA alters 16S rRNA processing. These findings allow KsgAs role in SSU biogenesis to be integrated into the network of other identified factors. Moreover, a methyltransferase-inactive form of KsgA, which we show to be deleterious to cell growth, profoundly impairs ribosome biogenesis-prompting discussion of KsgA as a possible antimicrobial drug target. These unexpected data suggest that methylation is a second layer of function for KsgA and that its critical role is as a supervisor of biogenesis of SSUs in vivo. These new findings and this proposed regulatory role offer a mechanistic explanation for the extreme conservation of the KsgA/Dim1p enzyme family.
Figures



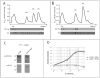
 ) or KsgA (
) or KsgA ( ) is overexpressed. Cells were grown to an OD600 of ∼0.5-0.6.
) is overexpressed. Cells were grown to an OD600 of ∼0.5-0.6.
Comment in
-
Ribosome biogenesis; the KsgA protein throws a methyl-mediated switch in ribosome assembly.Mol Microbiol. 2008 Dec;70(5):1051-3. doi: 10.1111/j.1365-2958.2008.06484.x. Mol Microbiol. 2008. PMID: 19006815
References
-
- Brimacombe R. The structure of ribosomal RNA: a three-dimensional jigsaw puzzle. Eur J Biochem. 1995;230:365–383. - PubMed
-
- Cunningham PR, Richard RB, Weitzmann CJ, Nurse K, Ofengand J. The absence of modified nucleotides affects both in vitro assembly and in vitro function of the 30S ribosomal subunit of Escherichia coli. Biochimie. 1991;73:789–796. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

