Temporal and functional profile of the transcriptional regulatory network in the early regenerative response to partial hepatectomy in the rat
- PMID: 18990226
- PMCID: PMC2613928
- DOI: 10.1186/1471-2164-9-527
Temporal and functional profile of the transcriptional regulatory network in the early regenerative response to partial hepatectomy in the rat
Abstract
Background: The goal of these studies was to characterize the transcriptional network regulating changes in gene expression in the remnant liver of the rat after 70% partial hepatectomy (PHx) during the early phase response including the transition of hepatocytes from the quiescent (G0) state and the onset of the G1 phase of the cell cycle.
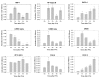
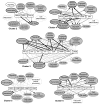
Results: The transcriptome of remnant livers was monitored at 1, 2, 4, and 6 hours after PHx using cDNA microarrays. Differentially regulated genes were grouped into six clusters according their temporal expression profiles. Promoter regions of genes in these clusters were examined for shared transcription factor binding sites (TFBS) by comparing enrichment of each TFBS relative to a reference set using the Promoter Analysis and Interaction Network Toolset (PAINT).Analysis of the gene expression time series data using ANOVA resulted in a total of 309 genes significantly up- or down-regulated at any of the four time points at a 20% FDR threshold. Sham-operated animals showed no significant differential expression. A subset of the differentially expressed genes was validated using quantitative RT-PCR. Distinct sets of TFBS could be identified that were significantly enriched in each one of the different temporal gene expression clusters. These included binding sites for transcription factors that had previously been recognized as contributing to the onset of regeneration, including NF-kappaB, C/EBP, HNF-1, CREB, as well as factors, such as ATF, AP-2, LEF-1, GATA and PAX-6, that had not yet been recognized to be involved in this process. A subset of these candidate TFBS was validated by measuring activation of corresponding transcription factors (HNF-1, NK-kappaB, CREB, C/EBP-alpha and C/EBP-beta, GATA-1, AP-2, PAX-6) in nuclear extracts from the remnant livers.
Conclusion: This analysis revealed multiple candidate transcription factors activated in the remnant livers, some known to be involved in the early phase of liver regeneration, and several not previously identified. The study describes the predominant temporal and functional elements to which these factors contribute and demonstrates the potential of this novel approach to define the functional correlates of the transcriptional regulatory network driving the early response to partial hepatectomy.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

