Genetic variation in tumor necrosis factor and the nuclear factor-kappaB canonical pathway and risk of non-Hodgkin's lymphoma
- PMID: 18990758
- PMCID: PMC2735864
- DOI: 10.1158/1055-9965.EPI-08-0536
Genetic variation in tumor necrosis factor and the nuclear factor-kappaB canonical pathway and risk of non-Hodgkin's lymphoma
Abstract
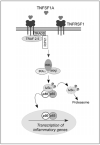
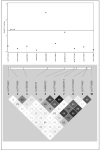
Non-Hodgkin's lymphoma (NHL) is a cancer closely associated with immune function, and the tumor necrosis factor (TNF) G-308A promoter polymorphism, which influences immune function and regulation, was recently reported by the InterLymph Consortium to be associated with NHL risk. TNF signaling activates the nuclear factor-kappaB (NF-kappaB) canonical pathway, leading to transcriptional activation of multiple genes that influence inflammation and immune response. We hypothesized that, in addition to TNF signaling, common genetic variation in genes from the NF-kappaB canonical pathway may affect risk of NHL. We genotyped 54 single nucleotide polymorphisms (SNP) within TNF, lymphotoxin A LTA, and nine NF-kappaB genes from the canonical pathway (TNFRSF1A, TRADD, TRAF2, TRAF5, RIPK1, CHUK, IKBKB, NFKB1, and REL) in a clinic-based study of 441 incident cases and 475 frequency-matched controls. Tagging SNPs were selected from HapMap supplemented by putative functional SNPs for LTA/TNF. We used principal components and haplo.stats to model gene-level associations and logistic regression to model SNP-level associations. Compared with the wild-type (GG), the AA genotype for the TNF promoter polymorphism G-308A (rs1800629) was associated with increased risk of NHL [odds ratio (OR), 2.14; 95% confidence interval (95% CI), 0.94-4.85], whereas the GA genotype was not (OR, 1.00; 95% CI, 0.74-1.34). This association was similar for follicular lymphoma and diffuse large B-cell lymphoma. A previously reported LTA/TNF haplotype was also associated with NHL risk. In gene-level analysis of the NF-kappaB pathway, only NFKB1 showed a statistically significant association with NHL (P = 0.049), and one NFKB1 tagSNP (rs4648022) was associated with NHL risk overall (ordinal OR, 0.59; 95% CI, 0.41-0.84; Ptrend = 0.0037) and for each of the common subtypes. In conclusion, we provide additional evidence for the role of genetic variation in TNF and LTA SNPs and haplotypes with risk of NHL and also provide some of the first preliminary evidence for an association of genetic variation in NFKB1, a downstream target of TNF signaling, with risk of NHL.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

