Gene expression profiling for molecular distinction and characterization of laser captured primary lung cancers
- PMID: 18992152
- PMCID: PMC2613386
- DOI: 10.1186/1479-5876-6-69
Gene expression profiling for molecular distinction and characterization of laser captured primary lung cancers
Abstract
Methods: We examined gene expression profiles of tumor cells from 29 untreated patients with lung cancer (10 adenocarcinomas (AC), 10 squamous cell carcinomas (SCC), and 9 small cell lung cancer (SCLC)) in comparison to 5 samples of normal lung tissue (NT). The European and American methodological quality guidelines for microarray experiments were followed, including the stipulated use of laser capture microdissection for separation and purification of the lung cancer tumor cells from surrounding tissue.
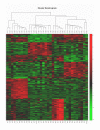
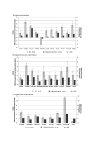
Results: Based on differentially expressed genes, different lung cancer samples could be distinguished from each other and from normal lung tissue using hierarchical clustering. Comparing AC, SCC and SCLC with NT, we found 205, 335 and 404 genes, respectively, that were at least 2-fold differentially expressed (estimated false discovery rate: < 2.6%). Different lung cancer subtypes had distinct molecular phenotypes, which also reflected their biological characteristics. Differentially expressed genes in human lung tumors which may be of relevance in the respective lung cancer subtypes were corroborated by quantitative real-time PCR. Genetic programming (GP) was performed to construct a classifier for distinguishing between AC, SCC, SCLC, and NT. Forty genes, that could be used to correctly classify the tumor or NT samples, have been identified. In addition, all samples from an independent test set of 13 further tumors (AC or SCC) were also correctly classified.
Conclusion: The data from this research identified potential candidate genes which could be used as the basis for the development of diagnostic tools and lung tumor type-specific targeted therapies.
Figures
References
-
- Sato M, Shames DS, Gazdar AF, Minna JD. A translational view of the molecular pathogenesis of lung cancer. J Thorac Oncol. 2007;2:327–43. - PubMed
-
- Hirabayashi H, Ohta M, Tanaka H, Sakaguchi M, Fujii Y, Miyoshi S, Matsuda H. Prognostic significance of p27KIP1 expression in resected non-small cell lung cancers: analysis in combination with expressions of p16INK4A, pRB, and p53. J Surg Oncol. 2002;81:177–84. doi: 10.1002/jso.10176. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

