Group I introns and associated homing endonuclease genes reveals a clinal structure for Porphyra spiralis var. amplifolia (Bangiales, Rhodophyta) along the Eastern coast of South America
- PMID: 18992156
- PMCID: PMC2585584
- DOI: 10.1186/1471-2148-8-308
Group I introns and associated homing endonuclease genes reveals a clinal structure for Porphyra spiralis var. amplifolia (Bangiales, Rhodophyta) along the Eastern coast of South America
Abstract
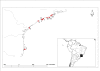
Background: Group I introns are found in the nuclear small subunit ribosomal RNA gene (SSU rDNA) of some species of the genus Porphyra (Bangiales, Rhodophyta). Size polymorphisms in group I introns has been interpreted as the result of the degeneration of homing endonuclease genes (HEG) inserted in peripheral loops of intron paired elements. In this study, intron size polymorphisms were characterized for different Porphyra spiralis var. amplifolia (PSA) populations on the Southern Brazilian coast, and were used to infer genetic relationships and genetic structure of these PSA populations, in addition to cox2-3 and rbcL-S regions. Introns of different sizes were tested qualitatively for in vitro self-splicing.
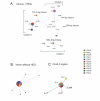
Results: Five intron size polymorphisms within 17 haplotypes were obtained from 80 individuals representing eight localities along the distribution of PSA in the Eastern coast of South America. In order to infer genetic structure and genetic relationships of PSA, these polymorphisms and haplotypes were used as markers for pairwise Fst analyses, Mantel's test and median joining network. The five cox2-3 haplotypes and the unique rbcL-S haplotype were used as markers for summary statistics, neutrality tests Tajima's D and Fu's Fs and for median joining network analyses. An event of demographic expansion from a population with low effective number, followed by a pattern of isolation by distance was obtained for PSA populations with the three analyses. In vitro experiments have shown that introns of different lengths were able to self-splice from pre-RNA transcripts.
Conclusion: The findings indicated that degenerated HEGs are reminiscent of the presence of a full-length and functional HEG, once fixed for PSA populations. The cline of HEG degeneration determined the pattern of isolation by distance. Analyses with the other markers indicated an event of demographic expansion from a population with low effective number. The different degrees of degeneration of the HEG do not refrain intron self-splicing. To our knowledge, this was the first study to address intraspecific evolutionary history of a nuclear group I intron; to use nuclear, mitochondrial and chloroplast DNA for population level analyses of Porphyra; and intron size polymorphism as a marker for population genetics.
Figures

Similar articles
-
The evolution of homing endonuclease genes and group I introns in nuclear rDNA.Mol Biol Evol. 2004 Jan;21(1):129-40. doi: 10.1093/molbev/msh005. Epub 2003 Oct 31. Mol Biol Evol. 2004. PMID: 14595099
-
The recent transfer of a homing endonuclease gene.Nucleic Acids Res. 2005 May 12;33(8):2734-41. doi: 10.1093/nar/gki564. Print 2005. Nucleic Acids Res. 2005. PMID: 15891115 Free PMC article.
-
Variant forms of a group I intron in nuclear small-subunit rRNA genes of the marine red alga Porphyra spiralis var. amplifolia.Mol Biol Evol. 1994 Mar;11(2):195-207. doi: 10.1093/oxfordjournals.molbev.a040102. Mol Biol Evol. 1994. PMID: 8170361
-
Invasion of a multitude of genetic niches by mobile endonuclease genes.FEMS Microbiol Lett. 2000 Apr 15;185(2):99-107. doi: 10.1111/j.1574-6968.2000.tb09046.x. FEMS Microbiol Lett. 2000. PMID: 10754232 Review.
-
Fungal mitochondrial genomes and genetic polymorphisms.Appl Microbiol Biotechnol. 2018 Nov;102(22):9433-9448. doi: 10.1007/s00253-018-9350-5. Epub 2018 Sep 12. Appl Microbiol Biotechnol. 2018. PMID: 30209549 Review.
Cited by
-
Horizontal transfer and gene conversion as an important driving force in shaping the landscape of mitochondrial introns.G3 (Bethesda). 2014 Apr 16;4(4):605-12. doi: 10.1534/g3.113.009910. G3 (Bethesda). 2014. PMID: 24515269 Free PMC article.
References
-
- Oliveira MC, Ragan MA. Variant forms of a group I intron in nuclear small-subunit rRNA genes of the marine red alga Porphyra spiralis var. amplifolia. Mol Biol Evol. 1994;11:195–207. - PubMed
-
- Müller KM, Cannone JJ, Gutell RR, Sheath RG. A Structural and Phylogenetic Analysis of the Group IC1 Introns in the Order Bangiales (Rhodophyta) Mol Biol Evol. 2001;18:1654–1667. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

