Efficient isolation of soluble intracellular single-chain antibodies using the twin-arginine translocation machinery
- PMID: 18992254
- PMCID: PMC2612092
- DOI: 10.1016/j.jmb.2008.10.051
Efficient isolation of soluble intracellular single-chain antibodies using the twin-arginine translocation machinery
Abstract
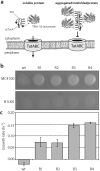
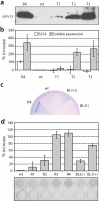
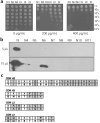
One of the most commonly used recombinant antibody formats is the single-chain variable fragment (scFv) that consists of the antibody variable heavy chain connected to the variable light chain by a flexible linker. Since disulfide bonds are often necessary for scFv folding, it can be challenging to express scFvs in the reducing environment of the cytosol. Thus, we sought to develop a method for antigen-independent selection of scFvs that are stable in the reducing cytosol of bacteria. To this end, we applied a recently developed genetic selection for protein folding and solubility based on the quality control feature of the Escherichia coli twin-arginine translocation (Tat) pathway. This selection employs a tripartite sandwich fusion of a protein-of-interest with an N-terminal Tat-specific signal peptide and C-terminal TEM1 beta-lactamase, thereby coupling antibiotic resistance with Tat pathway export. Here, we adapted this assay to develop intrabody selection after Tat export (ISELATE), a high-throughput selection strategy for the identification of solubility-enhanced scFv sequences. Using ISELATE for three rounds of laboratory evolution, it was possible to evolve a soluble scFv from an insoluble parental sequence. We show also that ISELATE enables focusing of an scFv library in soluble sequence space before functional screening and thus can be used to increase the likelihood of finding functional intrabodies. Finally, the technique was used to screen a large repertoire of naïve scFvs for clones that conferred significant levels of soluble accumulation. Our results reveal that the Tat quality control mechanism can be harnessed for molecular evolution of scFvs that are soluble in the reducing cytoplasm of E. coli.
Figures



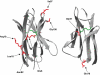
Similar articles
-
Genetic selection for protein solubility enabled by the folding quality control feature of the twin-arginine translocation pathway.Protein Sci. 2006 Mar;15(3):449-58. doi: 10.1110/ps.051902606. Epub 2006 Feb 1. Protein Sci. 2006. PMID: 16452624 Free PMC article.
-
Versatile selection technology for intracellular protein-protein interactions mediated by a unique bacterial hitchhiker transport mechanism.Proc Natl Acad Sci U S A. 2009 Mar 10;106(10):3692-7. doi: 10.1073/pnas.0704048106. Epub 2009 Feb 20. Proc Natl Acad Sci U S A. 2009. PMID: 19234130 Free PMC article.
-
Bacterial Inner-membrane Display for Screening a Library of Antibody Fragments.J Vis Exp. 2016 Oct 15;(116):54583. doi: 10.3791/54583. J Vis Exp. 2016. PMID: 27805609 Free PMC article.
-
Genetic selection of solubility-enhanced proteins using the twin-arginine translocation system.Methods Mol Biol. 2011;705:53-67. doi: 10.1007/978-1-61737-967-3_4. Methods Mol Biol. 2011. PMID: 21125380 Review.
-
Mechanistic Aspects of Folded Protein Transport by the Twin Arginine Translocase (Tat).J Biol Chem. 2015 Jul 3;290(27):16530-8. doi: 10.1074/jbc.R114.626820. Epub 2015 May 14. J Biol Chem. 2015. PMID: 25975269 Free PMC article. Review.
Cited by
-
Repurposing a bacterial quality control mechanism to enhance enzyme production in living cells.J Mol Biol. 2015 Mar 27;427(6 Pt B):1451-1463. doi: 10.1016/j.jmb.2015.01.003. Epub 2015 Jan 12. J Mol Biol. 2015. PMID: 25591491 Free PMC article.
-
An Intrabody Drug (rAAV6-INT41) Reduces the Binding of N-Terminal Huntingtin Fragment(s) to DNA to Basal Levels in PC12 Cells and Delays Cognitive Loss in the R6/2 Animal Model.J Neurodegener Dis. 2016;2016:7120753. doi: 10.1155/2016/7120753. Epub 2016 Aug 10. J Neurodegener Dis. 2016. PMID: 27595037 Free PMC article.
-
Directed evolution to improve protein folding in vivo.Curr Opin Struct Biol. 2018 Feb;48:117-123. doi: 10.1016/j.sbi.2017.12.003. Epub 2017 Dec 23. Curr Opin Struct Biol. 2018. PMID: 29278775 Free PMC article. Review.
-
Antibodies inside of a cell can change its outside: Can intrabodies provide a new therapeutic paradigm?Comput Struct Biotechnol J. 2016 Jul 31;14:304-8. doi: 10.1016/j.csbj.2016.07.003. eCollection 2016. Comput Struct Biotechnol J. 2016. PMID: 27570612 Free PMC article. Review.
-
Strain engineering for improved expression of recombinant proteins in bacteria.Microb Cell Fact. 2011 May 14;10:32. doi: 10.1186/1475-2859-10-32. Microb Cell Fact. 2011. PMID: 21569582 Free PMC article. Review.
References
-
- Holliger P, Hudson PJ. Engineered antibody fragments and the rise of single domains. Nat Biotechnol. 2005;23:1126–36. - PubMed
-
- Bird RE, Hardman KD, Jacobson JW, Johnson S, Kaufman BM, Lee SM, Lee T, Pope SH, Riordan GS, Whitlow M. Single-chain antigen-binding proteins. Science. 1988;242:423–6. - PubMed
-
- Huston JS, Levinson D, Mudgett-Hunter M, Tai MS, Novotny J, Margolies MN, Ridge RJ, Bruccoleri RE, Haber E, Crea R, et al. Protein engineering of antibody binding sites: recovery of specific activity in an anti-digoxin single-chain Fv analogue produced in Escherichia coli. Proc Natl Acad Sci U S A. 1988;85:5879–83. - PMC - PubMed
-
- Frisch C, Kolmar H, Schmidt A, Kleemann G, Reinhardt A, Pohl E, Uson I, Schneider TR, Fritz HJ. Contribution of the intramolecular disulfide bridge to the folding stability of REIv, the variable domain of a human immunoglobulin kappa light chain. Fold Des. 1996;1:431–40. - PubMed
-
- Kadokura H, Katzen F, Beckwith J. Protein disulfide bond formation in prokaryotes. Annu Rev Biochem. 2003;72:111–35. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

