ABF1-binding sites promote efficient global genome nucleotide excision repair
- PMID: 18996839
- PMCID: PMC3443742
- DOI: 10.1074/jbc.M806830200
ABF1-binding sites promote efficient global genome nucleotide excision repair
Abstract
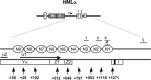
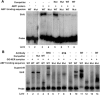
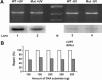
Global genome nucleotide excision repair (GG-NER) removes DNA damage from nontranscribing DNA. In Saccharomyces cerevisiae, the RAD7 and RAD16 genes are specifically required for GG-NER. We have reported that autonomously replicating sequence-binding factor 1 (ABF1) protein forms a stable complex with Rad7 and Rad16 proteins. ABF1 functions in transcription, replication, gene silencing, and NER in yeast. Here we show that binding of ABF1 to its DNA recognition sequence found at multiple genomic locations promotes efficient GG-NER in yeast. Mutation of the I silencer ABF1-binding site at the HMLalpha locus caused loss of ABF1 binding, which resulted in a domain of reduced GG-NER efficiency on one side of the ABF1-binding site. During GG-NER, nucleosome positioning at this site was not altered, and this correlated with an inability of the GG-NER complex to reposition nucleosomes in vitro.We discuss how the GG-NER complex might facilitate GG-NER while preventing unregulated gene transcription during this process.
Figures





Similar articles
-
Yeast autonomously replicating sequence binding factor is involved in nucleotide excision repair.Genes Dev. 1999 Dec 1;13(23):3052-8. doi: 10.1101/gad.13.23.3052. Genes Dev. 1999. PMID: 10601031 Free PMC article.
-
Functionally distinct nucleosome-free regions in yeast require Rad7 and Rad16 for nucleotide excision repair.DNA Repair (Amst). 2008 May 3;7(5):734-43. doi: 10.1016/j.dnarep.2008.01.016. Epub 2008 Mar 10. DNA Repair (Amst). 2008. PMID: 18329964
-
The yeast Rad7/Rad16/Abf1 complex generates superhelical torsion in DNA that is required for nucleotide excision repair.DNA Repair (Amst). 2004 Mar 4;3(3):277-87. doi: 10.1016/j.dnarep.2003.11.004. DNA Repair (Amst). 2004. PMID: 15177043
-
Nucleosome positioning, nucleotide excision repair and photoreactivation in Saccharomyces cerevisiae.DNA Repair (Amst). 2015 Dec;36:98-104. doi: 10.1016/j.dnarep.2015.09.012. Epub 2015 Sep 16. DNA Repair (Amst). 2015. PMID: 26429065 Review.
-
Histone modification and chromatin remodeling during NER.DNA Repair (Amst). 2015 Dec;36:105-113. doi: 10.1016/j.dnarep.2015.09.013. Epub 2015 Sep 16. DNA Repair (Amst). 2015. PMID: 26422133 Review.
Cited by
-
DNA repair mechanisms and the bypass of DNA damage in Saccharomyces cerevisiae.Genetics. 2013 Apr;193(4):1025-64. doi: 10.1534/genetics.112.145219. Genetics. 2013. PMID: 23547164 Free PMC article. Review.
-
Histone variant Htz1 promotes histone H3 acetylation to enhance nucleotide excision repair in Htz1 nucleosomes.Nucleic Acids Res. 2013 Oct;41(19):9006-19. doi: 10.1093/nar/gkt688. Epub 2013 Aug 7. Nucleic Acids Res. 2013. PMID: 23925126 Free PMC article.
-
How chromatin is remodelled during DNA repair of UV-induced DNA damage in Saccharomyces cerevisiae.PLoS Genet. 2011 Jun;7(6):e1002124. doi: 10.1371/journal.pgen.1002124. Epub 2011 Jun 16. PLoS Genet. 2011. PMID: 21698136 Free PMC article.
-
Rad7 E3 Ubiquitin Ligase Attenuates Polyubiquitylation of Rpn10 and Dsk2 Following DNA Damage in Saccharomyces cerevisiae.Adv Biol Chem. 2015;5(7):61944. doi: 10.4236/abc.2015.57021. Epub 2015 Dec 16. Adv Biol Chem. 2015. PMID: 27092291 Free PMC article.
-
DNA Repair in Haploid Context.Int J Mol Sci. 2021 Nov 17;22(22):12418. doi: 10.3390/ijms222212418. Int J Mol Sci. 2021. PMID: 34830299 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

