Allele-specific gene expression patterns in primary leukemic cells reveal regulation of gene expression by CpG site methylation
- PMID: 18997001
- PMCID: PMC2612957
- DOI: 10.1101/gr.083931.108
Allele-specific gene expression patterns in primary leukemic cells reveal regulation of gene expression by CpG site methylation
Abstract
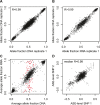
To identify genes that are regulated by cis-acting functional elements in acute lymphoblastic leukemia (ALL) we determined the allele-specific expression (ASE) levels of 2, 529 genes by genotyping a genome-wide panel of single nucleotide polymorphisms in RNA and DNA from bone marrow and blood samples of 197 children with ALL. Using a reproducible, quantitative genotyping method and stringent criteria for scoring ASE, we found that 16% of the analyzed genes display ASE in multiple ALL cell samples. For most of the genes, the level of ASE varied largely between the samples, from 1.4-fold overexpression of one allele to apparent monoallelic expression. For genes exhibiting ASE, 55% displayed bidirectional ASE in which overexpression of either of the two SNP alleles occurred. For bidirectional ASE we also observed overall higher levels of ASE and correlation with the methylation level of these sites. Our results demonstrate that CpG site methylation is one of the factors that regulates gene expression in ALL cells.
Figures




References
-
- Bray N.J., Buckland P.R., Owen M.J., O'Donovan M.C. Cis-acting variation in the expression of a high proportion of genes in human brain. Hum. Genet. 2003;113:149–153. - PubMed
-
- Carayon P., Portier M., Dussossoy D., Bord A., Petitpretre G., Canat X., Le Fur G., Casellas P. Involvement of peripheral benzodiazepine receptors in the protection of hematopoietic cells against oxygen radical damage. Blood. 1996;87:3170–3178. - PubMed
-
- Cheok M.H., Evans W.E. Acute lymphoblastic leukaemia: A model for the pharmacogenomics of cancer therapy. Nat. Rev. Cancer. 2006;6:117–129. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
