Rv0802c from Mycobacterium tuberculosis: the first structure of a succinyltransferase with the GNAT fold
- PMID: 18997321
- PMCID: PMC2581710
- DOI: 10.1107/S1744309108031679
Rv0802c from Mycobacterium tuberculosis: the first structure of a succinyltransferase with the GNAT fold
Abstract
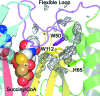
Gene rv0802c from Mycobacterium tuberculosis encodes a 218-amino-acid protein and is annotated as a hypothetical protein with homology to GCN5-related N-acetyltransferases. The structure of Rv0802c was determined in an unliganded form to 2.0 A resolution utilizing single-wavelength anomalous dispersion from a samarium soak that resulted in a single bound Sm(3+):citrate(2) complex. The structure confirms that Rv0802c exhibits the GCN5-related N-acetyltransferase fold and revealed a tetramer composed of a dimer of dimers with approximate 222 symmetry. In addition, a bound acetate ion indicated that Rv0802c may utilize a unique acyl donor for the family. The subsequent determination of the structure of Rv0802c in complex with succinyl-CoA to 2.3 A resolution suggests that Rv0802c is the first known GCN5-related N-acetyltransferase family member to utilize succinyl-CoA as a substrate.
Figures

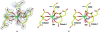


Similar articles
-
The crystal structure of Rv1347c, a putative antibiotic resistance protein from Mycobacterium tuberculosis, reveals a GCN5-related fold and suggests an alternative function in siderophore biosynthesis.J Biol Chem. 2005 Apr 8;280(14):13978-86. doi: 10.1074/jbc.M413904200. Epub 2005 Jan 28. J Biol Chem. 2005. PMID: 15695811
-
Rv0802c acetyltransferase from Mycobacterium tuberculosis H37Rv.Acta Microbiol Immunol Hung. 2005;52(3-4):363-71. doi: 10.1556/AMicr.52.2005.3-4.8. Acta Microbiol Immunol Hung. 2005. PMID: 16400876
-
Rv0802c is an acyltransferase that succinylates and acetylates Mycobacterium tuberculosis nucleoid-associated protein HU.Microbiology (Reading). 2021 Jul;167(7). doi: 10.1099/mic.0.001058. Microbiology (Reading). 2021. PMID: 34224344
-
The roles of bacterial GCN5-related N-acetyltransferases.Crit Rev Eukaryot Gene Expr. 2014;24(1):77-87. doi: 10.1615/critreveukaryotgeneexpr.2014007988. Crit Rev Eukaryot Gene Expr. 2014. PMID: 24579671 Review.
-
Structure and functions of the GNAT superfamily of acetyltransferases.Arch Biochem Biophys. 2005 Jan 1;433(1):212-26. doi: 10.1016/j.abb.2004.09.003. Arch Biochem Biophys. 2005. PMID: 15581578 Review.
Cited by
-
Bm-iAANAT3: Expression and characterization of a novel arylalkylamine N-acyltransferase from Bombyx mori.Arch Biochem Biophys. 2019 Jan;661:107-116. doi: 10.1016/j.abb.2018.11.015. Epub 2018 Nov 16. Arch Biochem Biophys. 2019. PMID: 30452894 Free PMC article.
-
Gcn5-Related N-Acetyltransferases (GNATs) With a Catalytic Serine Residue Can Play Ping-Pong Too.Front Mol Biosci. 2021 Apr 12;8:646046. doi: 10.3389/fmolb.2021.646046. eCollection 2021. Front Mol Biosci. 2021. PMID: 33912589 Free PMC article.
-
UniDrug-target: a computational tool to identify unique drug targets in pathogenic bacteria.PLoS One. 2012;7(3):e32833. doi: 10.1371/journal.pone.0032833. Epub 2012 Mar 14. PLoS One. 2012. PMID: 22431985 Free PMC article.
-
Toward Understanding the Essence of Post-Translational Modifications for the Mycobacterium tuberculosis Immunoproteome.Front Immunol. 2014 Aug 11;5:361. doi: 10.3389/fimmu.2014.00361. eCollection 2014. Front Immunol. 2014. PMID: 25157249 Free PMC article. Review.
-
The N-Acetylglutamate Synthase Family: Structures, Function and Mechanisms.Int J Mol Sci. 2015 Jun 9;16(6):13004-22. doi: 10.3390/ijms160613004. Int J Mol Sci. 2015. PMID: 26068232 Free PMC article. Review.
References
-
- Angus-Hill, M. L., Dutnall, R. N., Tafrov, S. T., Sternglanz, R. & Ramakrishnan, V. (1999). J. Mol. Biol.294, 1311–1325. - PubMed
-
- Card, G. L., Peterson, N. A., Smith, C. A., Rupp, B., Schick, B. M. & Baker, E. N. (2005). J. Biol. Chem.280, 13978–13986. - PubMed
-
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

