Structure of the Shigella T3SS effector IpaH defines a new class of E3 ubiquitin ligases
- PMID: 18997778
- PMCID: PMC2764551
- DOI: 10.1038/nsmb.1511
Structure of the Shigella T3SS effector IpaH defines a new class of E3 ubiquitin ligases
Abstract
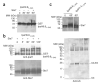
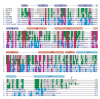
IpaH proteins are E3 ubiquitin ligases delivered by the type III secretion apparatus into host cells upon infection of humans by the Gram-negative pathogen Shigella flexneri. These proteins comprise a variable leucine-rich repeat-containing N-terminal domain and a conserved C-terminal domain harboring an invariant cysteine residue that is crucial for activity. IpaH homologs are encoded by diverse animal and plant pathogens. Here we demonstrate that the IpaH C-terminal domain carries the catalytic activity for ubiquitin transfer and that the N-terminal domain carries the substrate specificity. The structure of the IpaH C-terminal domain, determined to 2.65-A resolution, represents an all-helical fold bearing no resemblance to previously defined E3 ubiquitin ligases. The conserved and essential cysteine residue lies on a flexible, surface-exposed loop surrounded by conserved acidic residues, two of which are crucial for IpaH activity.
Figures


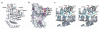


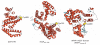
References
-
- Bhavsar AP, Guttman JA, Finlay BB. Manipulation of host-cell pathways by bacterial pathogens. Nature. 2007;449:827–834. - PubMed
-
- Galán JE, Cossart P. Host-pathogen interactions: a diversity of themes, a variety of molecular machines. Curr. Opin. Microbiol. 2005;8:1–3. - PubMed
-
- Ardley HC, Robinson PA. E3 ubiquitin ligases. Essays Biochem. 2005;41:15–30. - PubMed
-
- Mattoo S, Lee YM, Dixon JE. Interactions of bacterial effector proteins with host proteins. Curr. Opin. Immunol. 2007;19:392–401. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

