Structural basis for midbody targeting of spastin by the ESCRT-III protein CHMP1B
- PMID: 18997780
- PMCID: PMC2593743
- DOI: 10.1038/nsmb.1512
Structural basis for midbody targeting of spastin by the ESCRT-III protein CHMP1B
Abstract
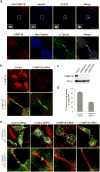
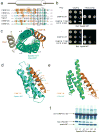
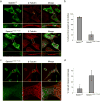
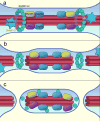
The endosomal sorting complex required for transport (ESCRT) machinery, including ESCRT-III, localizes to the midbody and participates in the membrane-abscission step of cytokinesis. The ESCRT-III protein charged multivesicular body protein 1B (CHMP1B) is required for recruitment of the MIT domain-containing protein spastin, a microtubule-severing enzyme, to the midbody. The 2.5-A structure of the C-terminal tail of CHMP1B with the MIT domain of spastin reveals a specific, high-affinity complex involving a noncanonical binding site between the first and third helices of the MIT domain. The structural interface is twice as large as that of the MIT domain of the VPS4-CHMP complex, consistent with the high affinity of the interaction. A series of unique hydrogen-bonding interactions and close packing of small side chains discriminate against the other ten human ESCRT-III subunits. Point mutants in the CHMP1B binding site of spastin block recruitment of spastin to the midbody and impair cytokinesis.
Figures


References
-
- Saksena S, Sun J, Chu T, Emr SD. ESCRTing proteins in the endocytic pathway. Trends Biochem Sci. 2007;32:561–573. - PubMed
-
- Morita E, Sundquist WI. Retrovirus budding. Ann Rev Cell Dev Biol. 2004;20:395–425. - PubMed
-
- Carlton JG, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 2007;316:1908–1912. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

