Genomic androgen receptor-occupied regions with different functions, defined by histone acetylation, coregulators and transcriptional capacity
- PMID: 18997859
- PMCID: PMC2577007
- DOI: 10.1371/journal.pone.0003645
Genomic androgen receptor-occupied regions with different functions, defined by histone acetylation, coregulators and transcriptional capacity
Abstract
Background: The androgen receptor (AR) is a steroid-activated transcription factor that binds at specific DNA locations and plays a key role in the etiology of prostate cancer. While numerous studies have identified a clear connection between AR binding and expression of target genes for a limited number of loci, high-throughput elucidation of these sites allows for a deeper understanding of the complexities of this process.
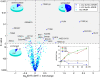
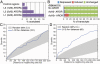
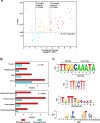
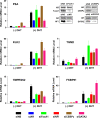
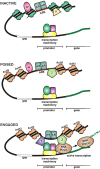
Methodology/principal findings: We have mapped 189 AR occupied regions (ARORs) and 1,388 histone H3 acetylation (AcH3) loci to a 3% continuous stretch of human genomic DNA using chromatin immunoprecipitation (ChIP) microarray analysis. Of 62 highly reproducible ARORs, 32 (52%) were also marked by AcH3. While the number of ARORs detected in prostate cancer cells exceeded the number of nearby DHT-responsive genes, the AcH3 mark defined a subclass of ARORs much more highly associated with such genes -- 12% of the genes flanking AcH3+ARORs were DHT-responsive, compared to only 1% of genes flanking AcH3-ARORs. Most ARORs contained enhancer activities as detected in luciferase reporter assays. Analysis of the AROR sequences, followed by site-directed ChIP, identified binding sites for AR transcriptional coregulators FoxA1, CEBPbeta, NFI and GATA2, which had diverse effects on endogenous AR target gene expression levels in siRNA knockout experiments.
Conclusions/significance: We suggest that only some ARORs function under the given physiological conditions, utilizing diverse mechanisms. This diversity points to differential regulation of gene expression by the same transcription factor related to the chromatin structure.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
- DK071122/DK/NIDDK NIH HHS/United States
- T32 CA009659/CA/NCI NIH HHS/United States
- C06 CA062528/CA/NCI NIH HHS/United States
- R01 LM008991/LM/NLM NIH HHS/United States
- R01 DK071122/DK/NIDDK NIH HHS/United States
- R01 CA109147/CA/NCI NIH HHS/United States
- RR14514-01/RR/NCRR NIH HHS/United States
- T32 CA009142/CA/NCI NIH HHS/United States
- P30 CA014089/CA/NCI NIH HHS/United States
- C06 RR10600-01/RR/NCRR NIH HHS/United States
- R56 DK071122/DK/NIDDK NIH HHS/United States
- C06 RR014514/RR/NCRR NIH HHS/United States
- P30 CA014089-30/CA/NCI NIH HHS/United States
- CA62528-01/CA/NCI NIH HHS/United States
- CA009142-26/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials

