Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome
- PMID: 18998772
- PMCID: PMC2581627
- DOI: 10.1371/journal.pbio.0060277
Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome
Abstract
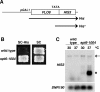
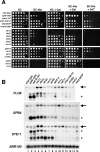
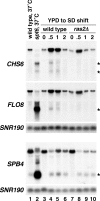
Previous studies in Saccharomyces cerevisiae have demonstrated that cryptic promoters within coding regions activate transcription in particular mutants. We have performed a comprehensive analysis of cryptic transcription in order to identify factors that normally repress cryptic promoters, to determine the amount of cryptic transcription genome-wide, and to study the potential for expression of genetic information by cryptic transcription. Our results show that a large number of factors that control chromatin structure and transcription are required to repress cryptic transcription from at least 1,000 locations across the S. cerevisiae genome. Two results suggest that some cryptic transcripts are translated. First, as expected, many cryptic transcripts contain an ATG and an open reading frame of at least 100 codons. Second, several cryptic transcripts are translated into proteins. Furthermore, a subset of cryptic transcripts tested is transiently induced in wild-type cells following a nutritional shift, suggesting a possible physiological role in response to a change in growth conditions. Taken together, our results demonstrate that, during normal growth, the global integrity of gene expression is maintained by a wide range of factors and suggest that, under altered genetic or physiological conditions, the expression of alternative genetic information may occur.
Conflict of interest statement
Figures



References
-
- Gingeras TR. Origin of phenotypes: genes and transcripts. Genome Res. 2007;17:682–690. - PubMed
-
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. - PubMed
-
- Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007;8:413–423. - PubMed
-
- Prasanth KV, Spector DL. Eukaryotic regulatory RNAs: an answer to the ‘genome complexity’ conundrum. Genes Dev. 2007;21:11–42. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

