Core structure of the yeast spt4-spt5 complex: a conserved module for regulation of transcription elongation
- PMID: 19000817
- PMCID: PMC2743916
- DOI: 10.1016/j.str.2008.08.013
Core structure of the yeast spt4-spt5 complex: a conserved module for regulation of transcription elongation
Abstract
The Spt4-Spt5 complex is an essential RNA polymerase II elongation factor found in all eukaryotes and important for gene regulation. We report here the crystal structure of Saccharomyces cerevisiae Spt4 bound to the NGN domain of Spt5. This structure reveals that Spt4-Spt5 binding is governed by an acid-dipole interaction between Spt5 and Spt4. Mutations that disrupt this interaction disrupt the complex. Residues forming this pivotal interaction are conserved in the archaeal homologs of Spt4 and Spt5, which we show also form a complex. Even though bacteria lack a Spt4 homolog, the NGN domains of Spt5 and its bacterial homologs are structurally similar. Spt4 is located at a position that may help to maintain the functional conformation of the following KOW domains in Spt5. This structural and evolutionary perspective of the Spt4-Spt5 complex and its homologs suggest that it is an ancient, core component of the transcription elongation machinery.
Figures

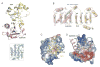




References
-
- Allison LA, Moyle M, Shales M, Ingles CJ. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell. 1985;42:599–610. - PubMed
-
- Artsimovitch I, Landick R. The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell. 2002;109:193–203. - PubMed
-
- Barton NH, Briggs DEG, Eisen JA, Goldstein DB, Patel NH. Evolution. Cold Spring Harbor Laboratory Press; 2007. Evolution; pp. 109–128.
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

