Atlas of gene expression in the developing kidney at microanatomic resolution
- PMID: 19000842
- PMCID: PMC2653061
- DOI: 10.1016/j.devcel.2008.09.007
Atlas of gene expression in the developing kidney at microanatomic resolution
Erratum in
- Dev Cell. 2009 Mar;16(3):482. Yu, Jing [added]
Abstract
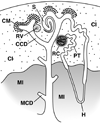
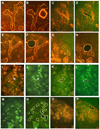
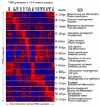
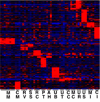
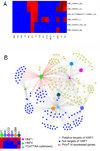
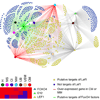
Kidney development is based on differential cell-type-specific expression of a vast number of genes. While multiple critical genes and pathways have been elucidated, a genome-wide analysis of gene expression within individual cellular and anatomic structures is lacking. Accomplishing this could provide significant new insights into fundamental developmental mechanisms such as mesenchymal-epithelial transition, inductive signaling, branching morphogenesis, and segmentation. We describe here a comprehensive gene expression atlas of the developing mouse kidney based on the isolation of each major compartment by either laser capture microdissection or fluorescence-activated cell sorting, followed by microarray profiling. The resulting data agree with known expression patterns and additional in situ hybridizations. This kidney atlas allows a comprehensive analysis of the progression of gene expression states during nephrogenesis, as well as discovery of potential growth factor-receptor interactions. In addition, the results provide deeper insight into the genetic regulatory mechanisms of kidney development.
Figures

References
-
- Akpinar P, Kuwajima S, Krutzfeldt J, Stoffel M. Tmem27: a cleaved and shed plasma membrane protein that stimulates pancreatic beta cell proliferation. [see comment] Cell Metabolism. 2005;2:385–397. - PubMed
-
- Almeida M, Han L, Martin-Millan M, O'Brien CA, Manolagas SC. Oxidative stress antagonizes Wnt signaling in osteoblast precursors by diverting beta-catenin from T cell factor- to forkhead box O-mediated transcription. J Biol Chem. 2007;282:27298–27305. - PubMed
-
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. 1995;57:289–300.
-
- Boyle S, Misfeldt A, Chandler KJ, Deal KK, Southard-Smith EM, Mortlock DP, Baldwin HS, de Caestecker M. Fate mapping using Cited1-CreERT2 mice demonstrates that the cap mesenchyme contains self-renewing progenitor cells and gives rise exclusively to nephronic epithelia. Developmental Biology. 2008;313:234–245. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

