Transcription factor Fli1 regulates collagen fibrillogenesis in mouse skin
- PMID: 19001092
- PMCID: PMC2612518
- DOI: 10.1128/MCB.01278-08
Transcription factor Fli1 regulates collagen fibrillogenesis in mouse skin
Abstract
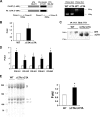
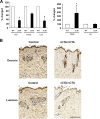
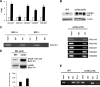
Biosynthesis of fibrillar collagen in the skin is precisely regulated to maintain proper tissue homeostasis; however, the molecular mechanisms involved in this process remain largely unknown. Transcription factor Fli1 has been shown to repress collagen synthesis in cultured dermal fibroblasts. This study investigated the role of Fli1 in regulation of collagen biosynthesis in mice skin in vivo using mice with the homozygous deletion of the C-terminal transcriptional activation (CTA) domain of the Fli1 gene (Fli1(DeltaCTA/DeltaCTA)). Skin analyses of the Fli1 mutant mice revealed a significant upregulation of fibrillar collagen genes at mRNA level, as well as increased collagen content as measured by acetic acid extraction and hydroxyproline assays. In addition, collagen fibrils contained ultrastructural abnormalities including immature thin fibrils and very thick irregularly shaped fibrils, which correlated with the reduced levels of decorin, fibromodulin, and lumican. Fibroblasts cultured from the skin of Fli1(DeltaCTA/DeltaCTA) mice maintained elevated synthesis of collagen mRNA and protein. Additional experiments in cultured fibroblasts have revealed that although Fli1 DeltaCTA retains the ability to bind to the collagen promoter in vitro and in vivo, it no longer functions as transcriptional repressor. Together, these results establish Fli1 as a key regulator of the collagen homeostasis in the skin in vivo.
Figures

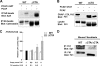
References
-
- Arvand, A., S. M. Welford, M. A. Teitell, and C. T. Denny. 2001. The COOH-terminal domain of FLI-1 is necessary for full tumorigenesis and transcriptional modulation by EWS/FLI-1. Cancer Res. 615311-5317. - PubMed
-
- Asano, Y., J. Czuwara-Ladykowska, and M. Trojanowska. 2007. Transforming growth factor-beta regulates DNA binding activity of transcription factor Fli1 by p300/CREB-binding protein-associated factor-dependent acetylation. J. Biol. Chem. 28234672-34683. - PubMed
-
- Canty, E. G., and K. E. Kadler. 2005. Procollagen trafficking, processing and fibrillogenesis. J. Cell Sci. 1181341-1353. - PubMed
-
- Chakravarti, S. 2002. Functions of lumican and fibromodulin: lessons from knockout mice. Glycoconj. J. 19287-293. - PubMed
-
- Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162156-159. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
