The H4 tail domain participates in intra- and internucleosome interactions with protein and DNA during folding and oligomerization of nucleosome arrays
- PMID: 19001093
- PMCID: PMC2612503
- DOI: 10.1128/MCB.01343-08
The H4 tail domain participates in intra- and internucleosome interactions with protein and DNA during folding and oligomerization of nucleosome arrays
Abstract
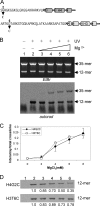
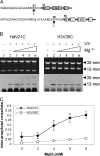
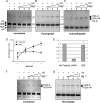
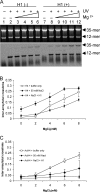
The condensation of nucleosome arrays into higher-order secondary and tertiary chromatin structures likely involves long-range internucleosomal interactions mediated by the core histone tail domains. We have characterized interarray interactions mediated by the H4 tail domain, known to play a predominant role in the formation of such structures. We find that the N-terminal end of the H4 tail mediates interarray contacts with DNA during self-association of oligonucleosome arrays similar to that found previously for the H3 tail domain. However, a site near the histone fold domain of H4 participates in a distinct set of interactions, contacting both DNA and H2A in condensed structures. Moreover, we also find that H4-H2A interactions occur via an intra- as well as an internucleosomal fashion, supporting an additional intranucleosomal function for the tail. Interestingly, acetylation of the H4 tail has little effect on interarray interactions by itself but overrides the strong stimulation of interarray interactions induced by linker histones. Our results indicate that the H4 tail facilitates secondary and tertiary chromatin structure formation via a complex array of potentially exclusive interactions that are distinct from those of the H3 tail domain.
Figures

References
-
- Annunziato, A. A., L.-L. Y. Frado, R. L. Seale, and C. L. F. Woodcock. 1988. Treatment with sodium butyrate inhibits the complete condensation of interphase chromatin. Chromosoma 96132-138. - PubMed
-
- Ausio, J., F. Dong, and K. E. van Holde. 1989. Use of selectively trypsinized nucleosome core particles to analyze the role of the histone “tails” in the stabilization of the nucleosome. J. Mol. Biol. 206451-463. - PubMed
-
- Barski, A., S. Cuddapah, K. Cui, T. Y. Roh, D. E. Schones, Z. Wang, G. Wei, I. Chepelev, and K. Zhao. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129823-837. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
