Escherichia coli tmRNA lacking pseudoknot 1 tags truncated proteins in vivo and in vitro
- PMID: 19001120
- PMCID: PMC2612775
- DOI: 10.1261/rna.1192409
Escherichia coli tmRNA lacking pseudoknot 1 tags truncated proteins in vivo and in vitro
Abstract
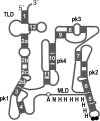
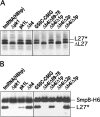
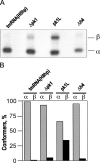
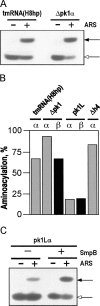
Transfer-messenger RNA (tmRNA) and protein SmpB facilitate trans-translation, a quality-control process that tags truncated proteins with short peptides recognized by a number of proteases and recycles ribosomes stalled at the 3' end of mRNA templates lacking stop codons. The tmRNA molecule is a hybrid of tRNA- and mRNA-like domains that are usually connected by four pseudoknots (pk1-pk4). Replacement of pk1 with a single-stranded RNA yields pk1L, a mutant tmRNA that tags truncated proteins very poorly in vitro but very efficiently in vivo. However, deletion of the whole pk1 is deleterious for protein tagging. In contrast, deletion of helix 4 yields Deltah4, a fully functional tmRNA derivative containing a single hairpin instead of pk1. Further deletions in the pk1 segment yield two subclasses of mutant tmRNAs that are unable to tag truncated proteins, but some of them bind to stalled ribosomes. Our studies demonstrate that pk1 is not essential for tmRNA functions but contributes to the stability of the tmRNA structure. Our studies also indicate that the length of this RNA segment is critical for both tmRNA binding to the ribosome and resumption of translation.
Figures

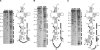
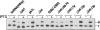


References
-
- Barends S., Wower J., Kraal B. Kinetic parameters for tmRNA binding to alanyl-tRNA synthetase and elongation factor Tu from Escherichia coli . Biochemistry. 2000;39:2652–2658. - PubMed
-
- Barends S., Karzai A.W., Sauer R.T., Wower J., Kraal B. Simultaneous and functional binding of SmpB and EF-Tu-TP to the alanyl acceptor arm of tmRNA. J. Mol. Biol. 2001;314:9–21. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
