Histone H3 lysine 56 acetylation by Rtt109 is crucial for chromosome positioning
- PMID: 19001125
- PMCID: PMC2582893
- DOI: 10.1083/jcb.200806065
Histone H3 lysine 56 acetylation by Rtt109 is crucial for chromosome positioning
Abstract
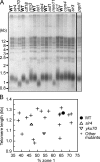
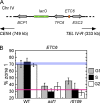
Correct intranuclear organization of chromosomes is crucial for many genome functions, but the mechanisms that position chromatin are not well understood. We used a layered screen to identify Saccharomyces cerevisiae mutants defective in telomere localization to the nuclear periphery. We find that events in S phase are crucial for correct telomere localization. In particular, the histone chaperone Asf1 functions in telomere peripheral positioning. Asf1 stimulates acetylation of histone H3 lysine 56 (H3K56) by the histone acetyltransferase Rtt109. Analysis of rtt109Delta and H3K56 mutants suggests that the acetylation/deacetylation cycle of the H3K56 residue is required for proper telomere localization. The function of H3K56 acetylation in localizing chromosome domains is not confined to telomeres because deletion of RTT109 also prevents the correct peripheral localization of a newly identified S. cerevisiae "chromosome-organizing clamp" locus. Because chromosome positioning is subject to epigenetic inheritance, H3K56 acetylation may mediate correct chromosome localization by facilitating accurate transmission of chromatin status during DNA replication.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

