Pds1p is required for meiotic recombination and prophase I progression in Saccharomyces cerevisiae
- PMID: 19001291
- PMCID: PMC2621190
- DOI: 10.1534/genetics.108.095513
Pds1p is required for meiotic recombination and prophase I progression in Saccharomyces cerevisiae
Abstract
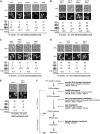
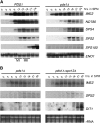
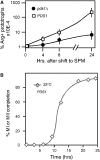
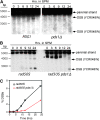
Sister-chromatid separation at the metaphase-anaphase transition is regulated by a proteolytic cascade. Destruction of the securin Pds1p liberates the Esp1p separase, which ultimately targets the mitotic cohesin Mcd1p/Scc1p for destruction. Pds1p stabilization by the spindle or DNA damage checkpoints prevents sister-chromatid separation while mutants lacking PDS1 (pds1Delta) are temperature sensitive for growth due to elevated chromosome loss. This report examined the role of the budding yeast Pds1p in meiotic progression using genetic, cytological, and biochemical assays. Similar to its mitotic function, Pds1p destruction is required for metaphase I-anaphase I transition. However, even at the permissive temperature for growth, pds1Delta mutants arrest with prophase I spindle and nuclear characteristics. This arrest was partially suppressed by preventing recombination initiation or by inactivating a subset of recombination checkpoint components. Further studies revealed that Pds1p is required for recombination in both double-strand-break formation and synaptonemal complex assembly. Although deleting PDS1 did not affect the degradation of the meiotic cohesin Rec8p, Mcd1p was precociously destroyed as cells entered the meiotic program. This role is meiosis specific as Mcd1p destruction is not altered in vegetative pds1Delta cultures. These results define a previously undescribed role for Pds1p in cohesin maintenance, recombination, and meiotic progression.
Figures




Similar articles
-
An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast.Cell. 1998 Jun 12;93(6):1067-76. doi: 10.1016/s0092-8674(00)81211-8. Cell. 1998. PMID: 9635435
-
Recombination protein Tid1p controls resolution of cohesin-dependent linkages in meiosis in Saccharomyces cerevisiae.J Cell Biol. 2005 Oct 24;171(2):241-53. doi: 10.1083/jcb.200505020. Epub 2005 Oct 17. J Cell Biol. 2005. PMID: 16230461 Free PMC article.
-
Pds1p is required for faithful execution of anaphase in the yeast, Saccharomyces cerevisiae.J Cell Biol. 1996 Apr;133(1):85-97. doi: 10.1083/jcb.133.1.85. J Cell Biol. 1996. PMID: 8601616 Free PMC article.
-
Meiotic prophase-like pathway for cleavage-independent removal of cohesin for chromosome morphogenesis.Curr Genet. 2019 Aug;65(4):817-827. doi: 10.1007/s00294-019-00959-x. Epub 2019 Mar 28. Curr Genet. 2019. PMID: 30923890 Review.
-
Cell cycle mechanisms of sister chromatid separation; roles of Cut1/separin and Cut2/securin.Genes Cells. 2000 Jan;5(1):1-8. doi: 10.1046/j.1365-2443.2000.00306.x. Genes Cells. 2000. PMID: 10651900 Review.
Cited by
-
Recipes and mechanisms of cellular reprogramming: a case study on budding yeast Saccharomyces cerevisiae.BMC Syst Biol. 2011 Apr 12;5:50. doi: 10.1186/1752-0509-5-50. BMC Syst Biol. 2011. PMID: 21486480 Free PMC article.
-
Meiotic control of the APC/C: similarities & differences from mitosis.Cell Div. 2011 Aug 1;6:16. doi: 10.1186/1747-1028-6-16. Cell Div. 2011. PMID: 21806783 Free PMC article.
-
Proximity labeling reveals new functional relationships between meiotic recombination proteins in S. cerevisiae.PLoS Genet. 2024 Oct 15;20(10):e1011432. doi: 10.1371/journal.pgen.1011432. eCollection 2024 Oct. PLoS Genet. 2024. PMID: 39405359 Free PMC article.
-
Mutually dependent degradation of Ama1p and Cdc20p terminates APC/C ubiquitin ligase activity at the completion of meiotic development in yeast.Cell Div. 2013 Jul 1;8(1):9. doi: 10.1186/1747-1028-8-9. Cell Div. 2013. PMID: 23816140 Free PMC article.
-
Ama1p-activated anaphase-promoting complex regulates the destruction of Cdc20p during meiosis II.Mol Biol Cell. 2011 Feb 1;22(3):315-26. doi: 10.1091/mbc.E10-04-0360. Epub 2010 Nov 30. Mol Biol Cell. 2011. PMID: 21118994 Free PMC article.
References
-
- Alani, E., R. Padmore and N. Kleckner, 1990. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell 61 419–436. - PubMed
-
- Arora, C., K. Kee, S. Maleki and S. Keeney, 2004. Antiviral protein Ski8 is a direct partner of Spo11 in meiotic DNA break formation, independent of its cytoplasmic role in RNA metabolism. Mol. Cell 13 549–559. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

