How does arrestin assemble MAPKs into a signaling complex?
- PMID: 19001375
- PMCID: PMC2610502
- DOI: 10.1074/jbc.M806124200
How does arrestin assemble MAPKs into a signaling complex?
Abstract
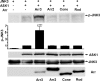
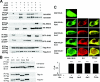
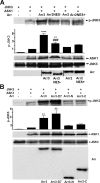
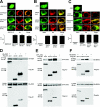
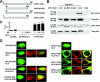
Arrestins bind active phosphorylated G protein-coupled receptors, precluding G protein activation and channeling signaling to alternative pathways. Arrestins also function as mitogen-activated protein kinase (MAPK) scaffolds, bringing together three components of MAPK signaling modules. Here we have demonstrated that all four vertebrate arrestins interact with JNK3, MKK4, and ASK1, but only arrestin3 facilitates JNK3 activation. Thus, the functional specificity of arrestins is not determined by differential binding of the kinases. Using receptor binding-impaired mutant, we have shown that free arrestin3 readily promotes JNK3 phosphorylation. We identified key arrestin-binding elements in JNK3 and ASK1 and investigated the molecular interactions of arrestin2 and arrestin3 and their individual domains with the components of the two MAPK cascades, ASK1-MKK4-JNK3 and c-Raf-1-MEK1-ERK2. We found that both arrestin domains interact with all six kinases. These findings shed new light on the mechanism of arrestin-mediated MAPK activation and the spatial arrangement of the three kinases on arrestin molecule.
Figures



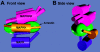
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

