The leukemogenic t(8;21) fusion protein AML1-ETO controls rRNA genes and associates with nucleolar-organizing regions at mitotic chromosomes
- PMID: 19001502
- PMCID: PMC2904240
- DOI: 10.1242/jcs.033431
The leukemogenic t(8;21) fusion protein AML1-ETO controls rRNA genes and associates with nucleolar-organizing regions at mitotic chromosomes
Abstract
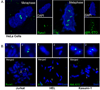
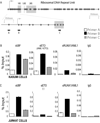
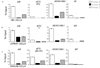
RUNX1/AML1 is required for definitive hematopoiesis and is frequently targeted by chromosomal translocations in acute myeloid leukemia (AML). The t(8;21)-related AML1-ETO fusion protein blocks differentiation of myeloid progenitors. Here, we show by immunofluorescence microscopy that during interphase, endogenous AML1-ETO localizes to nuclear microenvironments distinct from those containing native RUNX1/AML1 protein. At mitosis, we clearly detect binding of AML1-ETO to nucleolar-organizing regions in AML-derived Kasumi-1 cells and binding of RUNX1/AML1 to the same regions in Jurkat cells. Both RUNX1/AML1 and AML1-ETO occupy ribosomal DNA repeats during interphase, as well as interact with the endogenous RNA Pol I transcription factor UBF1. Promoter cytosine methylation analysis indicates that RUNX1/AML1 binds to rDNA repeats that are more highly CpG methylated than those bound by AML1-ETO. Downregulation by RNA interference reveals that RUNX1/AML1 negatively regulates rDNA transcription, whereas AML1-ETO is a positive regulator in Kasumi-1 cells. Taken together, our findings identify a novel role for the leukemia-related AML1-ETO protein in epigenetic control of cell growth through upregulation of ribosomal gene transcription mediated by RNA Pol I, consistent with the hyper-proliferative phenotype of myeloid cells in AML patients.
Figures





References
-
- Bernardin-Fried F, Kummalue T, Leijen S, Collector MI, Ravid K, Friedman AD. AML1/RUNX1 increases during G1 to S cell cycle progression independent of cytokine-dependent phosphorylation and induces cyclin D3 gene expression. J. Biol. Chem. 2004;279:15678–15687. - PubMed
-
- Budde A, Grummt I. p53 represses ribosomal gene transcription. Oncogene. 1999;18:1119–1124. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

