KDM2A represses transcription of centromeric satellite repeats and maintains the heterochromatic state
- PMID: 19001877
- PMCID: PMC2636745
- DOI: 10.4161/cc.7.22.7062
KDM2A represses transcription of centromeric satellite repeats and maintains the heterochromatic state
Abstract
Heterochromatin plays an essential role in the preservation of epigenetic information, the transcriptional repression of repetitive DNA elements and inactive genes, and the proper segregation of chromosomes during mitosis. Here we identify KDM2A, a JmjC-domain containing histone demethylase, as a heterochromatin-associated and HP1-interacting protein that promotes HP1 localization to chromatin. We show that KDM2A is required to maintain the heterochromatic state, as determined using a candidate-based approach coupled to an in vivo epigenetic reporter system. Remarkably, a parallel and independent siRNA screen also detected a role for KDM2A in epigenetic silencing. Moreover, we demonstrate that KDM2A associates with centromeres and represses transcription of small non-coding RNAs that are encoded by the clusters of satellite repeats at the centromere. Dissecting the relationship between heterochromatin and centromeric RNA transcription is the basis of ongoing studies. We demonstrate that forced expression of these satellite RNA transcripts compromise the heterochromatic state and HP1 localization to chromatin. Finally, we show that KDM2A is required to sustain centromeric integrity and genomic stability, particularly during mitosis. Since the disruption of epigenetic control mechanisms contributes to cellular transformation, these results, together with the low levels of KDM2A found in prostate carcinomas, suggest a role for KDM2A in cancer development.
Figures
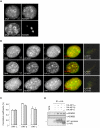
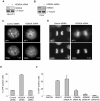

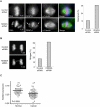
Comment in
-
Heterochromatin: lost in transcription?Cell Cycle. 2008 Nov 15;7(22):3479-80. doi: 10.4161/cc.7.22.7321. Epub 2008 Nov 15. Cell Cycle. 2008. PMID: 19001850 No abstract available.
Similar articles
-
KDM2A integrates DNA and histone modification signals through a CXXC/PHD module and direct interaction with HP1.Nucleic Acids Res. 2017 Feb 17;45(3):1114-1129. doi: 10.1093/nar/gkw979. Nucleic Acids Res. 2017. PMID: 28180290 Free PMC article.
-
The chromatin remodelling complex NoRC safeguards genome stability by heterochromatin formation at telomeres and centromeres.EMBO Rep. 2013 Aug;14(8):704-10. doi: 10.1038/embor.2013.87. Epub 2013 Jun 25. EMBO Rep. 2013. PMID: 23797874 Free PMC article.
-
JmjC enzyme KDM2A is a regulator of rRNA transcription in response to starvation.EMBO J. 2010 May 5;29(9):1510-22. doi: 10.1038/emboj.2010.56. Epub 2010 Apr 8. EMBO J. 2010. PMID: 20379134 Free PMC article.
-
Satellite DNAs between selfishness and functionality: structure, genomics and evolution of tandem repeats in centromeric (hetero)chromatin.Gene. 2008 Feb 15;409(1-2):72-82. doi: 10.1016/j.gene.2007.11.013. Epub 2007 Dec 4. Gene. 2008. PMID: 18182173 Review.
-
Studies on the mechanism of RNAi-dependent heterochromatin assembly.Cold Spring Harb Symp Quant Biol. 2006;71:461-71. doi: 10.1101/sqb.2006.71.044. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381328 Review.
Cited by
-
Transcriptional repression of histone deacetylase 3 by the histone demethylase KDM2A is coupled to tumorigenicity of lung cancer cells.J Biol Chem. 2014 Mar 14;289(11):7483-96. doi: 10.1074/jbc.M113.521625. Epub 2014 Jan 30. J Biol Chem. 2014. PMID: 24482232 Free PMC article.
-
A demethylation deficient isoform of the lysine demethylase KDM2A interacts with pericentromeric heterochromatin in an HP1a-dependent manner.Nucleus. 2017 Sep 3;8(5):563-572. doi: 10.1080/19491034.2017.1342915. Epub 2017 Aug 17. Nucleus. 2017. PMID: 28816576 Free PMC article.
-
De novo variants in KDM2A cause a syndromic neurodevelopmental disorder.medRxiv [Preprint]. 2025 Apr 1:2025.03.31.25324695. doi: 10.1101/2025.03.31.25324695. medRxiv. 2025. PMID: 40236430 Free PMC article. Preprint.
-
Chromatin and oxygen sensing in the context of JmjC histone demethylases.Biochem J. 2014 Sep 15;462(3):385-95. doi: 10.1042/BJ20140754. Biochem J. 2014. PMID: 25145438 Free PMC article. Review.
-
Identification of protein complexes that bind to histone H3 combinatorial modifications using super-SILAC and weighted correlation network analysis.Nucleic Acids Res. 2015 Feb 18;43(3):1418-32. doi: 10.1093/nar/gku1350. Epub 2015 Jan 20. Nucleic Acids Res. 2015. PMID: 25605797 Free PMC article.
References
-
- Maison C, Almouzni G. HP1 and the dynamics of heterochromatin maintenance. Nat Rev Mol Cell Biol. 2004;5:296–304. - PubMed
-
- Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. - PubMed
-
- Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, Chen T, Li E, Jenuwein T, Peters AH. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol. 2003;13:1192–200. - PubMed
-
- Peters AH, O'Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, Opravil S, Doyle M, Sibilia M, Jenuwein T. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–37. - PubMed
-
- Hall IM, Shankaranarayana GD, Noma K, Ayoub N, Cohen A, Grewal SI. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
