Stem cell marking with promotor-deprived self-inactivating retroviral vectors does not lead to induced clonal imbalance
- PMID: 19002163
- PMCID: PMC2834973
- DOI: 10.1038/mt.2008.238
Stem cell marking with promotor-deprived self-inactivating retroviral vectors does not lead to induced clonal imbalance
Abstract
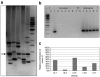
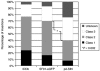
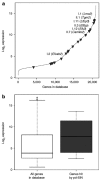
Stable genetic modification of stem cells holds great promise for gene therapy and marking, but commonly used gamma-retroviral vectors were found to influence growth/survival characteristics of hematopoietic stem cells (HSCs) by insertional mutagenesis. In this article, we show that promoter-deprived gamma-retroviral self-inactivating (pd-SIN) vectors allow stable genetic marking of serially reconstituting murine HSC. In contrast to findings with gamma-retroviral long terminal repeat (LTR) vectors, serial transplantation of pd-SIN-marked HSC in a sensitive mouse model was apparently not associated with induced clonal imbalance of gene-marked HSC. Furthermore, insertions of pd-SIN into protooncogenes, growth-promoting and signaling genes occurred significantly less frequent than in control experiments with LTR vectors. Also, transcriptional dysregulation of neighboring genes potentially caused by the pd-SIN insertion was rarely seen and comparatively weak. The integration pattern of promotor-deprived SIN vectors in reconstituting HSC seems to depend on the transcriptional activity of the respective gene loci reflecting the picture described for LTR vectors. In conclusion, our data strongly support the use of SIN vectors for gene-marking studies and suggest an increased therapeutic index for vectors lacking enhancers active in HSC.
Figures



Similar articles
-
Comparative clonal analysis of reconstitution kinetics after transplantation of hematopoietic stem cells gene marked with a lentiviral SIN or a γ-retroviral LTR vector.Exp Hematol. 2013 Jan;41(1):28-38.e3. doi: 10.1016/j.exphem.2012.09.003. Epub 2012 Sep 16. Exp Hematol. 2013. PMID: 22989760
-
Clonal dominance of hematopoietic stem cells triggered by retroviral gene marking.Science. 2005 May 20;308(5725):1171-4. doi: 10.1126/science.1105063. Science. 2005. PMID: 15905401
-
Cell-culture assays reveal the importance of retroviral vector design for insertional genotoxicity.Blood. 2006 Oct 15;108(8):2545-53. doi: 10.1182/blood-2005-08-024976. Epub 2006 Jul 6. Blood. 2006. PMID: 16825499 Free PMC article.
-
Insertional mutagenesis and clonal dominance: biological and statistical considerations.Gene Ther. 2008 Jan;15(2):143-53. doi: 10.1038/sj.gt.3303052. Epub 2007 Nov 1. Gene Ther. 2008. PMID: 17972922 Review.
-
Retroviral Insertional Mutagenesis in Humans: Evidence for Four Genetic Mechanisms Promoting Expansion of Cell Clones.Mol Ther. 2020 Feb 5;28(2):352-356. doi: 10.1016/j.ymthe.2019.12.009. Epub 2020 Jan 7. Mol Ther. 2020. PMID: 31951833 Free PMC article. Review.
Cited by
-
Clonal Dominance With Retroviral Vector Insertions Near the ANGPT1 and ANGPT2 Genes in a Human Xenotransplant Mouse Model.Mol Ther Nucleic Acids. 2014 Oct 7;3(10):e200. doi: 10.1038/mtna.2014.51. Mol Ther Nucleic Acids. 2014. PMID: 25291142 Free PMC article.
-
Gene therapy for hemophilia.Front Biosci (Landmark Ed). 2015 Jan 1;20(3):556-603. doi: 10.2741/4324. Front Biosci (Landmark Ed). 2015. PMID: 25553466 Free PMC article. Review.
-
Clonal competition in BcrAbl-driven leukemia: how transplantations can accelerate clonal conversion.Mol Cancer. 2017 Jul 14;16(1):120. doi: 10.1186/s12943-017-0668-x. Mol Cancer. 2017. PMID: 28709463 Free PMC article.
-
Radiation rescue: mesenchymal stromal cells protect from lethal irradiation.PLoS One. 2011 Jan 5;6(1):e14486. doi: 10.1371/journal.pone.0014486. PLoS One. 2011. PMID: 21245929 Free PMC article.
-
Self-inactivating retroviral vector-mediated gene transfer induces oncogene activation and immortalization of primary murine bone marrow cells.Mol Ther. 2009 Nov;17(11):1910-8. doi: 10.1038/mt.2009.172. Epub 2009 Jul 28. Mol Ther. 2009. PMID: 19638958 Free PMC article.
References
-
- Dick JE, Magli MC, Huszar D, Phillips RA., and , Bernstein A. Introduction of a selectable gene into primitive stem cells capable of long-term reconstitution of the hemopoietic system of W/Wv mice. Cell. 1985;42:71–79. - PubMed
-
- Lemischka IR, Raulet DH., and , Mulligan RC. Developmental potential and dynamic behavior of hematopoietic stem cells. Cell. 1986;45:917–927. - PubMed
-
- Tey SK., and , Brenner MK. The continuing contribution of gene marking to cell and gene therapy. Mol Ther. 2007;15:666–676. - PubMed
-
- Hacein-Bey-Abina S, Le Deist F, Carlier F, Bouneaud C, Hue C, De Villartay JP, et al. Sustained correction of X-linked severe combined immunodeficiency by ex vivo gene therapy. N Engl J Med. 2002;346:1185–1193. - PubMed
-
- Aiuti A, Slavin S, Aker M, Ficara F, Deola S, Mortellaro A, et al. Correction of ADA-SCID by stem cell gene therapy combined with nonmyeloablative conditioning. Science. 2002;296:2410–2413. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

