Expanding or restricting the target site repertoire of zinc-finger nucleases: the inter-domain linker as a major determinant of target site selectivity
- PMID: 19002164
- PMCID: PMC2834978
- DOI: 10.1038/mt.2008.233
Expanding or restricting the target site repertoire of zinc-finger nucleases: the inter-domain linker as a major determinant of target site selectivity
Abstract
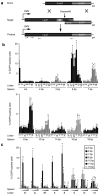
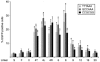
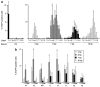
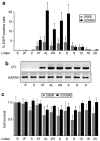
Precise manipulations of complex genomes by zinc-finger nucleases (ZFNs) depend on site-specific DNA cleavage, which requires two ZFN subunits to bind to two target half-sites separated by a spacer of 6 base pairs (bp). ZFN subunits consist of a specific DNA-binding domain and a nonspecific cleavage domain, connected by a short inter-domain linker. In this study, we conducted a systematic analysis of 11 candidate-based linkers using episomal and chromosomal targets in two human cell lines. We achieved gene targeting in up to 20% of transfected cells and identified linker variants that enforce DNA cleavage at narrowly defined spacer lengths and linkers that expand the repertoire of potential target sites. For instance, a nine amino acid (aa) linker induced efficient gene conversion at chromosomal sites with 7- or 16-bp spacers, whereas 4-aa linkers had activity optima at 5- and 6-bp spacers. Notably, single aa substitutions in the 4-aa linker affected the ZFN activity significantly, and both gene conversion and ZFN-associated toxicity depended on the linker/spacer combination and the cell type. In summary, both sequence and length of the inter-domain linker determine ZFN activity and target-site specificity, and are therefore important parameters to account for when designing ZFNs for genome editing.
Figures

References
-
- Cathomen T., and , Keith Joung J. Zinc-finger nucleases: the next generation emerges. Mol Ther. 2008;16:1200–1207. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

