One step at a time: endoplasmic reticulum-associated degradation
- PMID: 19002207
- PMCID: PMC2654601
- DOI: 10.1038/nrm2546
One step at a time: endoplasmic reticulum-associated degradation
Abstract
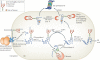
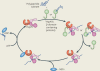
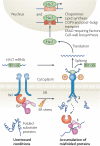
Protein folding in the endoplasmic reticulum (ER) is monitored by ER quality control (ERQC) mechanisms. Proteins that pass ERQC criteria traffic to their final destinations through the secretory pathway, whereas non-native and unassembled subunits of multimeric proteins are degraded by the ER-associated degradation (ERAD) pathway. During ERAD, molecular chaperones and associated factors recognize and target substrates for retrotranslocation to the cytoplasm, where they are degraded by the ubiquitin-proteasome machinery. The discovery of diseases that are associated with ERAD substrates highlights the importance of this pathway. Here, we summarize our current understanding of each step during ERAD, with emphasis on the factors that catalyse distinct activities.
Figures
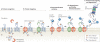
References
-
- Ellis RJ. Macromolecular crowding: obvious but underappreciated. Trends Biochem. Sci. 2001;26:597–604. - PubMed
-
- Despa F, Orgill DP, Lee RC. Molecular crowding effects on protein stability. Ann. NY Acad. Sci. 2005;1066:54–66. - PubMed
-
- Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. - PubMed
-
- Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

