Evolving synaptic plasticity with an evolutionary cellular development model
- PMID: 19002249
- PMCID: PMC2577323
- DOI: 10.1371/journal.pone.0003697
Evolving synaptic plasticity with an evolutionary cellular development model
Abstract
Since synaptic plasticity is regarded as a potential mechanism for memory formation and learning, there is growing interest in the study of its underlying mechanisms. Recently several evolutionary models of cellular development have been presented, but none have been shown to be able to evolve a range of biological synaptic plasticity regimes. In this paper we present a biologically plausible evolutionary cellular development model and test its ability to evolve different biological synaptic plasticity regimes. The core of the model is a genomic and proteomic regulation network which controls cells and their neurites in a 2D environment. The model has previously been shown to successfully evolve behaving organisms, enable gene related phenomena, and produce biological neural mechanisms such as temporal representations. Several experiments are described in which the model evolves different synaptic plasticity regimes using a direct fitness function. Other experiments examine the ability of the model to evolve simple plasticity regimes in a task -based fitness function environment. These results suggest that such evolutionary cellular development models have the potential to be used as a research tool for investigating the evolutionary aspects of synaptic plasticity and at the same time can serve as the basis for novel artificial computational systems.
Conflict of interest statement
Figures



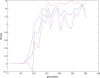
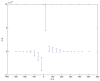
 . Spike time measures are given in system epochs, synaptic change Δwt is given in absolute synaptic weight values. Each plot presents the average synaptic change values from 24 sessions of 1000 epochs. The error bars are set at one standard deviation.
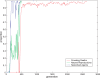
. Spike time measures are given in system epochs, synaptic change Δwt is given in absolute synaptic weight values. Each plot presents the average synaptic change values from 24 sessions of 1000 epochs. The error bars are set at one standard deviation.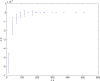
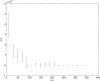
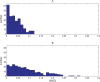
 . Spike time measures are given in system epochs; synaptic change Δwt is given in absolute synaptic weight values. Each plot presents the average synaptic change values from 24 sessions of 1000 epochs. The error bars are set at one standard deviation.
. Spike time measures are given in system epochs; synaptic change Δwt is given in absolute synaptic weight values. Each plot presents the average synaptic change values from 24 sessions of 1000 epochs. The error bars are set at one standard deviation.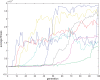

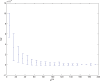
 (defined as the time interval since the last pre-synaptic spike), taken from a 20th generation virtual organism evolved in a session with fitness function
(defined as the time interval since the last pre-synaptic spike), taken from a 20th generation virtual organism evolved in a session with fitness function  . The post-synaptic cell was set as non-active during measurements. Spike time measures are given in system epochs; synaptic change Δwt is given in absolute synaptic weight values. Each plot presents the average synaptic change values from 24 sessions of 1000 epochs. The error bars are of one standard deviation.
. The post-synaptic cell was set as non-active during measurements. Spike time measures are given in system epochs; synaptic change Δwt is given in absolute synaptic weight values. Each plot presents the average synaptic change values from 24 sessions of 1000 epochs. The error bars are of one standard deviation.
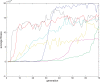
 (defined as the time interval since the last post-synaptic spike), taken from a 20th generation virtual organism evolved in a session with fitness function
(defined as the time interval since the last post-synaptic spike), taken from a 20th generation virtual organism evolved in a session with fitness function  . The pre-synaptic cell was set as non -active during measurements. Spike time measures are given in system epochs; synaptic change Δwt is given in absolute synaptic weight values. Each plot presents the average synaptic change values from 24 sessions of 1000 epochs. The error bars are set atone standard deviation.
. The pre-synaptic cell was set as non -active during measurements. Spike time measures are given in system epochs; synaptic change Δwt is given in absolute synaptic weight values. Each plot presents the average synaptic change values from 24 sessions of 1000 epochs. The error bars are set atone standard deviation.
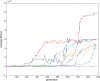
References
-
- Montana D, Lawrance D. Training Feed Forward networks using genetic algorithms. 1989. Proc. of Int. Joint Conf. on Artificial Intelligence; Detroit, Michigan, USA.
-
- Harp S, Samad T, Guha A. Toward the genetic synthesis of neural networks. In: S DJ, editor. Third Intl. Conf. on Genetic Algorithms; Virginia, USA: Morgan Kaufmann; 1989. pp. 360–369.
-
- Rust AG. Developmental self Organization in Artificial Neural Networks. 1998. Ph.D: University of Hertfordshire.
-
- Rust AG, Adams R, Bolouri H. Evolutionary neural topiary: Growing and sculpting artificial neurons to order. In: B MA, M JS, P NH, R S, editors. Artificial Life VII: The 7th International Conference. Cambridge, MA: MIT Press; 2000.
-
- Kitano H. Designing neural networks using genetic algorithms with graph generation system. Complex Systems. 1990;4:461–476.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

