The yeast mitochondrial citrate transport protein: identification of the Lysine residues responsible for inhibition mediated by Pyridoxal 5'-phosphate
- PMID: 19002576
- PMCID: PMC2775541
- DOI: 10.1007/s10863-008-9187-1
The yeast mitochondrial citrate transport protein: identification of the Lysine residues responsible for inhibition mediated by Pyridoxal 5'-phosphate
Abstract
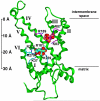
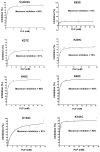
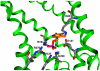
The present investigation identifies the molecular basis for the well-documented inhibition of the mitochondrial inner membrane citrate transport protein (CTP) function by the lysine-selective reagent pyridoxal 5'-phosphate. Kinetic analysis indicates that PLP is a linear mixed inhibitor of the Cys-less CTP, with a predominantly competitive component. We have previously concluded that the CTP contains at least two substrate binding sites which are located at increasing depths within the substrate translocation pathway and which contain key lysine residues. In the present investigation, the roles of Lys-83 in substrate binding site one, Lys-37 and Lys-239 in substrate binding site two, and four other off-pathway lysines in conferring PLP-inhibition of transport was determined by functional characterization of seven lysine to cysteine substitution mutants. We observed that replacement of Lys-83 with cysteine resulted in a 78% loss of the PLP-mediated inhibition of CTP function. In contrast, replacement of either Lys-37 or Lys-239 with cysteine caused a modest reduction in the inhibition caused by PLP (i.e., 31% and 20% loss of inhibition, respectively). Interestingly, these losses of PLP-mediated inhibition could be rescued by covalent modification of each cysteine with MTSEA, a reagent that adds a lysine-like moiety (i.e. SCH(2)CH(2)NH(3) (+)) to the cysteine sulfhydryl group. Importantly, the replacement of non-binding site lysines (i.e., Lys-45, Lys-48, Lys-134, Lys-141) with cysteine resulted in little change in the PLP inhibition. Based upon these results, we conducted docking calculations with the CTP structural model leading to the development of a physical binding model for PLP. In combination, our data support the conclusion that PLP exerts its main inhibitory effect by binding to residues located within the two substrate binding sites of the CTP, with Lys-83 being the primary determinant of the total PLP effect since the replacement of this single lysine abolishes nearly all of the observed inhibition by PLP.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

